CEBRA: a self-supervised learning algorithm for obtaining interpretable, Consistent EmBeddings of high-dimensional Recordings using Auxiliary variables
CEBRA is a machine-learning method that can be used to compress time series in a way that reveals otherwise hidden structures in the variability of the data. It excels on behavioural and neural data recorded simultaneously. We have shown it can be used to decode the activity from the visual cortex of the mouse brain to reconstruct a viewed video, to decode trajectories from the sensoirmotor cortex of primates, and for decoding position during navigation. For these use cases and other demos see our Documentation.
Demo Applications
Application of CEBRA-Behavior to rat hippocampus data (Grosmark and Buzsáki, 2016), showing position/neural activity (left), overlayed with decoding obtained by CEBRA. The current point in embedding space is highlighted (right). CEBRA obtains a median absolute error of 5cm (total track length: 160cm; see Schneider et al. 2023 for details). Video is played at 2x real-time speed.
Interactive visualization of the CEBRA embedding for the rat hippocampus data. This 3D plot shows how neural activity is mapped to a lower-dimensional space that correlates with the animal's position and movement direction. Open In Colaboratory
CEBRA applied to mouse primary visual cortex, collected at the Allen Institute (de Vries et al. 2020, Siegle et al. 2021). 2-photon and Neuropixels recordings are embedded with CEBRA using DINO frame features as labels. The embedding is used to decode the video frames using a kNN decoder on the CEBRA-Behavior embedding from the test set.
CEBRA applied to M1 and S1 neural data, demonstrating how neural activity from primary motor and somatosensory cortices can be effectively embedded and analyzed. See DeWolf et al. 2024 for details.
Publications
Steffen Schneider*, Jin Hwa Lee*, Mackenzie Weygandt Mathis. Nature 2023
A comprehensive introduction to CEBRA, demonstrating its capabilities in joint behavioral and neural analysis across various datasets and species.
Read Paper PreprintSteffen Schneider, Rodrigo González Laiz, Anastasiia Filipova, Markus Frey, Mackenzie Weygandt Mathis. AISTATS 2025
An extension of CEBRA that provides attribution maps for time-series data using regularized contrastive learning.
Read Paper Preprint NeurIPS-W 2023 VersionPatent Information
Please note EPFL has filed a patent titled "Dimensionality reduction of time-series data, and systems and devices that use the resultant embeddings" so if this does not work for your non-academic use case, please contact the Tech Transfer Office at EPFL.
Overview
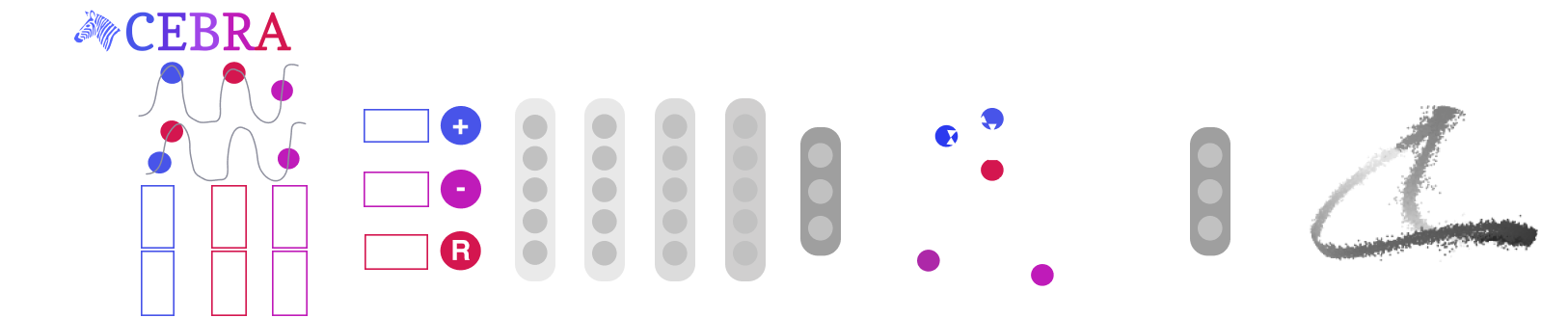
Mapping behavioural actions to neural activity is a fundamental goal of neuroscience. As our ability to record large neural and behavioural data increases, there is growing interest in modeling neural dynamics during adaptive behaviors to probe neural representations. In particular, neural latent embeddings can reveal underlying correlates of behavior, yet, we lack non-linear techniques that can explicitly and flexibly leverage joint behavior and neural data to uncover neural dynamics. Here, we fill this gap with a novel encoding method, CEBRA, that jointly uses behavioural and neural data in a (supervised) hypothesis- or (self-supervised) discovery-driven manner to produce both consistent and high-performance latent spaces. We show that consistency can be used as a metric for uncovering meaningful differences, and the inferred latents can be used for decoding. We validate its accuracy and demonstrate our tool's utility for both calcium and electrophysiology datasets, across sensory and motor tasks, and in simple or complex behaviors across species. It allows for single and multi-session datasets to be leveraged for hypothesis testing or can be used label-free. Lastly, we show that CEBRA can be used for the mapping of space, uncovering complex kinematic features, produces consistent latent spaces across 2-photon and Neuropixels data, and can provide rapid, high-accuracy decoding of natural movies from visual cortex.
Software
You can find our official implementation of the CEBRA algorithm on GitHub: Watch and Star the repository to be notified of future updates and releases. You can also follow us on Twitter for updates on the project.
If you are interested in collaborations, please contact us via email.
BibTeX
Please cite our papers as follows:
author={Steffen Schneider and Jin Hwa Lee and Mackenzie Weygandt Mathis},
title={Learnable latent embeddings for joint behavioural and neural analysis},
journal={Nature},
year={2023},
month={May},
day={03},
issn={1476-4687},
doi={10.1038/s41586-023-06031-6},
url={https://doi.org/10.1038/s41586-023-06031-6}
}
title={Time-series attribution maps with regularized contrastive learning},
author={Steffen Schneider and Rodrigo Gonz{\'a}lez Laiz and Anastasiia Filippova and Markus Frey and Mackenzie Weygandt Mathis},
booktitle={The 28th International Conference on Artificial Intelligence and Statistics},
year={2025},
url={https://proceedings.mlr.press/v258/schneider25a.html}
}
Impact & Citations
CEBRA has been cited in numerous high-impact publications across neuroscience, machine learning, and related fields. Our work has influenced research in neural decoding, brain-computer interfaces, computational neuroscience, and machine learning methods for time-series analysis.
Our research has been cited in proceedings and journals including Nature Science ICML Nature Neuroscience ICML Neuron NeurIPS ICLR and others.
