Technical: Training models across animals#
This notebook will demo how to use a multisession CEBRA implementation.
We will compare embeddings obtained on 4 rat datasets when trained on four single-session models vs. one multisession model.
We will use CEBRA-Behavior (
conditional='time_delta') for both single and multi session implementation.Each individual single session output embedding is aligned to the first session.
How multi-session training works:
For flexibility, it is implemented so that it does not fit one model for all sessions. Consequently, it is possible to use sessions that do not have the same number of data features (e.g., not the same number of neurons from one session to the other). The number of samples can also vary (true for single session too).
It fits one model per session but the pos/neg sampling is performed across all sessions, making the models invariant to the labels across all sessions.
When to use multi-session training:
Check out the following list to verify if our multisession implementation is the right tool for your needs.
I have multiple sessions or animals that I want to consider as a pseudo-subject and use them jointly for training CEBRA.
That is the case because of limited access to simultaneously recorded neurons or looking for animal-invariant features in the neural data.
I want to get more consistent embeddings from one session or animal to the other.
I want to be able to use CEBRA for a new session that is fully unseen during training (e.g., could be useful for a brain machine interface applications).
WARNING: I am not interested in the influence of individual variations of the label features from one session/animal to the other on the resulting embedding. I do not need those session or animal specific information.
Install note
Be sure you have cebra, and the demo dependencies, installed to use this notebook:
[1]:
!pip install --pre 'cebra[datasets,demos]'
[2]:
import sys
import matplotlib.pyplot as plt
import cebra.data
import cebra.datasets
import cebra.integrations
from cebra import CEBRA
import matplotlib.pyplot as plt
import pickle
Load the data#
The data will be automatically downloaded into a
/datafolder.
[2]:
hippocampus_a = cebra.datasets.init('rat-hippocampus-single-achilles')
hippocampus_b = cebra.datasets.init('rat-hippocampus-single-buddy')
hippocampus_c = cebra.datasets.init('rat-hippocampus-single-cicero')
hippocampus_g = cebra.datasets.init('rat-hippocampus-single-gatsby')
100%|██████████| 10.0M/10.0M [00:00<00:00, 10.6MB/s]
Download complete. Dataset saved in 'data/rat_hippocampus/achilles.jl'
100%|██████████| 2.68M/2.68M [00:00<00:00, 7.83MB/s]
Download complete. Dataset saved in 'data/rat_hippocampus/buddy.jl'
100%|██████████| 22.0M/22.0M [00:01<00:00, 12.2MB/s]
Download complete. Dataset saved in 'data/rat_hippocampus/cicero.jl'
100%|██████████| 9.40M/9.40M [00:01<00:00, 8.92MB/s]
Download complete. Dataset saved in 'data/rat_hippocampus/gatsby.jl'
[5]:
names = ["achilles", "buddy", "cicero", "gatsby"]
datas = [hippocampus_a.neural.numpy(), hippocampus_b.neural.numpy(), hippocampus_c.neural.numpy(), hippocampus_g.neural.numpy()]
labels = [hippocampus_a.continuous_index.numpy(), hippocampus_b.continuous_index.numpy(), hippocampus_c.continuous_index.numpy(), hippocampus_g.continuous_index.numpy()]
Single-session training#
Create and fit one single session model per dataset
[6]:
max_iterations = 15000 # by default, 5000 iterations
[7]:
embeddings = dict()
# Single session training
for name, X, y in zip(names, datas, labels):
# Fit one CEBRA model per session (i.e., per rat)
print(f"Fitting CEBRA for {name}")
cebra_model = CEBRA(model_architecture='offset10-model',
batch_size=512,
learning_rate=3e-4,
temperature=1,
output_dimension=3,
max_iterations=max_iterations,
distance='cosine',
conditional='time_delta',
device='cuda_if_available',
verbose=True,
time_offsets=10)
cebra_model.fit(X, y)
embeddings[name] = cebra_model.transform(X)
# Align the single session embeddings to the first rat
alignment = cebra.data.helper.OrthogonalProcrustesAlignment()
first_rat = list(embeddings.keys())[0]
for j, rat_name in enumerate(list(embeddings.keys())[1:]):
embeddings[f"{rat_name}"] = alignment.fit_transform(
embeddings[first_rat], embeddings[rat_name], labels[0], labels[j+1])
# Save embeddings in current folder
with open('embeddings.pkl', 'wb') as f:
pickle.dump(embeddings, f)
Fitting CEBRA for achilles
pos: 0.0999 neg: 5.4138 total: 5.5137 temperature: 1.0000: 100%|██████████████████████████████████████████████████████████████| 15000/15000 [02:49<00:00, 88.47it/s]
Fitting CEBRA for buddy
pos: 0.1868 neg: 5.4300 total: 5.6168 temperature: 1.0000: 100%|██████████████████████████████████████████████████████████████| 15000/15000 [02:51<00:00, 87.46it/s]
Fitting CEBRA for cicero
pos: 0.2931 neg: 5.4676 total: 5.7607 temperature: 1.0000: 100%|██████████████████████████████████████████████████████████████| 15000/15000 [02:54<00:00, 85.85it/s]
Fitting CEBRA for gatsby
pos: 0.1416 neg: 5.4502 total: 5.5918 temperature: 1.0000: 100%|██████████████████████████████████████████████████████████████| 15000/15000 [02:52<00:00, 87.13it/s]
Multisession training#
Create and fit one multi-session model on all datasets
[8]:
multi_embeddings = dict()
# Multisession training
multi_cebra_model = CEBRA(model_architecture='offset10-model',
batch_size=512,
learning_rate=3e-4,
temperature=1,
output_dimension=3,
max_iterations=max_iterations,
distance='cosine',
conditional='time_delta',
device='cuda_if_available',
verbose=True,
time_offsets=10)
# Provide a list of data, i.e. datas = [data_a, data_b, ...]
multi_cebra_model.fit(datas, labels)
# Transform each session with the right model, by providing the corresponding session ID
for i, (name, X) in enumerate(zip(names, datas)):
multi_embeddings[name] = multi_cebra_model.transform(X, session_id=i)
# Save embeddings in current folder
with open('multi_embeddings.pkl', 'wb') as f:
pickle.dump(multi_embeddings, f)
pos: 0.1853 neg: 6.8037 total: 6.9890 temperature: 1.0000: 100%|██████████████████████████████████████████████████████████████| 15000/15000 [09:38<00:00, 25.92it/s]
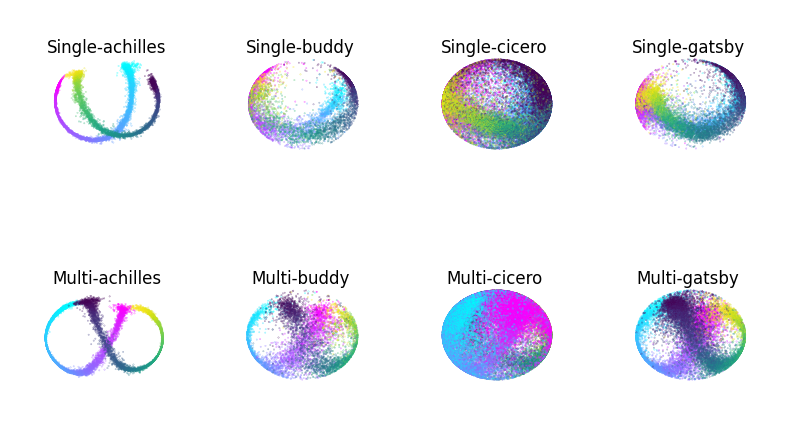
Compare embeddings#
Also see Extended Data Figure 7 of Schneider, Lee, Mathis.
[9]:
with open('embeddings.pkl', 'rb') as f:
embeddings = pickle.load(f)
with open('multi_embeddings.pkl', 'rb') as f:
multi_embeddings = pickle.load(f)
[10]:
def plot_hippocampus(ax, embedding, label, gray = False, idx_order = (0,1,2)):
r_ind = label[:,1] == 1
l_ind = label[:,2] == 1
if not gray:
r_cmap = 'cool'
l_cmap = 'viridis'
r_c = label[r_ind, 0]
l_c = label[l_ind, 0]
else:
r_cmap = None
l_cmap = None
r_c = 'gray'
l_c = 'gray'
idx1, idx2, idx3 = idx_order
r=ax.scatter(embedding [r_ind,idx1],
embedding [r_ind,idx2],
embedding [r_ind,idx3],
c=r_c,
cmap=r_cmap, s=0.05, alpha=0.75)
l=ax.scatter(embedding [l_ind,idx1],
embedding [l_ind,idx2],
embedding [l_ind,idx3],
c=l_c,
cmap=l_cmap, s=0.05, alpha=0.75)
ax.grid(False)
ax.xaxis.pane.fill = False
ax.yaxis.pane.fill = False
ax.zaxis.pane.fill = False
ax.xaxis.pane.set_edgecolor('w')
ax.yaxis.pane.set_edgecolor('w')
ax.zaxis.pane.set_edgecolor('w')
return ax
Note to Google Colaboratory users: replace the first line of the next cell (%matplotlib notebook) with %matplotlib inline.
[11]:
%matplotlib notebook
fig = plt.figure(figsize=(10,6))
ax1 = plt.subplot(241, projection='3d')
ax2 = plt.subplot(242, projection='3d')
ax3 = plt.subplot(243, projection='3d')
ax4 = plt.subplot(244, projection='3d')
axs = [ax1, ax2, ax3, ax4]
ax5 = plt.subplot(245, projection='3d')
ax6 = plt.subplot(246, projection='3d')
ax7 = plt.subplot(247, projection='3d')
ax8 = plt.subplot(248, projection='3d')
axs_multi = [ax5, ax6, ax7, ax8]
for name, label, ax, ax_multi in zip(names, labels, axs, axs_multi):
ax = plot_hippocampus(ax, embeddings[name], label)
ax.set_title(f'Single-{name}', y=1, pad=-20)
ax.axis('off')
ax_multi = plot_hippocampus(ax_multi, multi_embeddings[name], label)
ax_multi.set_title(f'Multi-{name}', y=1, pad=-20)
ax_multi.axis('off')
plt.subplots_adjust(wspace=0,
hspace=0)
plt.show()
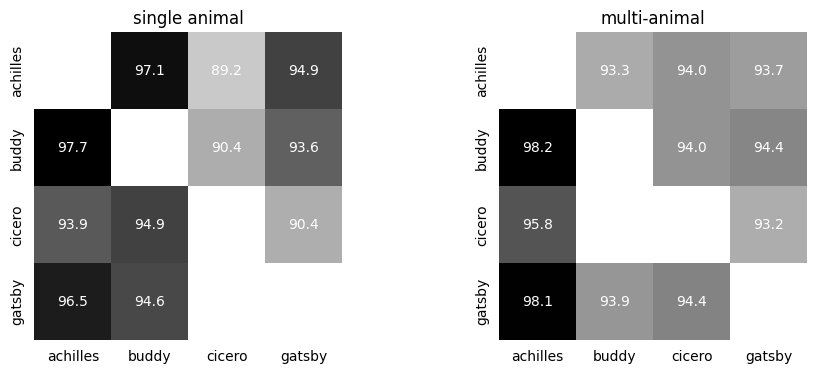
Compute the consistency maps#
[12]:
# labels to align the subjects is the position of the mouse in the arena
labels = [data.continuous_index[:, 0]
for data in [hippocampus_a, hippocampus_b, hippocampus_c, hippocampus_g]]
# CEBRA (single animal)
scores, pairs, subjects = cebra.sklearn.metrics.consistency_score(embeddings=list(embeddings.values()),
labels=labels,
dataset_ids=names,
between="datasets")
# CEBRA (multiple animals)
multi_scores, multi_pairs, multi_subjects = cebra.sklearn.metrics.consistency_score(embeddings=list(multi_embeddings.values()),
labels=labels,
dataset_ids=names,
between="datasets")
[13]:
%matplotlib notebook
# Display consistency maps
fig = plt.figure(figsize=(11, 4))
ax1 = plt.subplot(121)
ax2 = plt.subplot(122)
ax1 = cebra.plot_consistency(scores, pairs=pairs, datasets=subjects,
ax=ax1, title="single animal", colorbar_label=None)
ax2 = cebra.plot_consistency(multi_scores, pairs=multi_pairs, datasets=multi_subjects,
ax=ax2, title="multi-animal", colorbar_label=None)