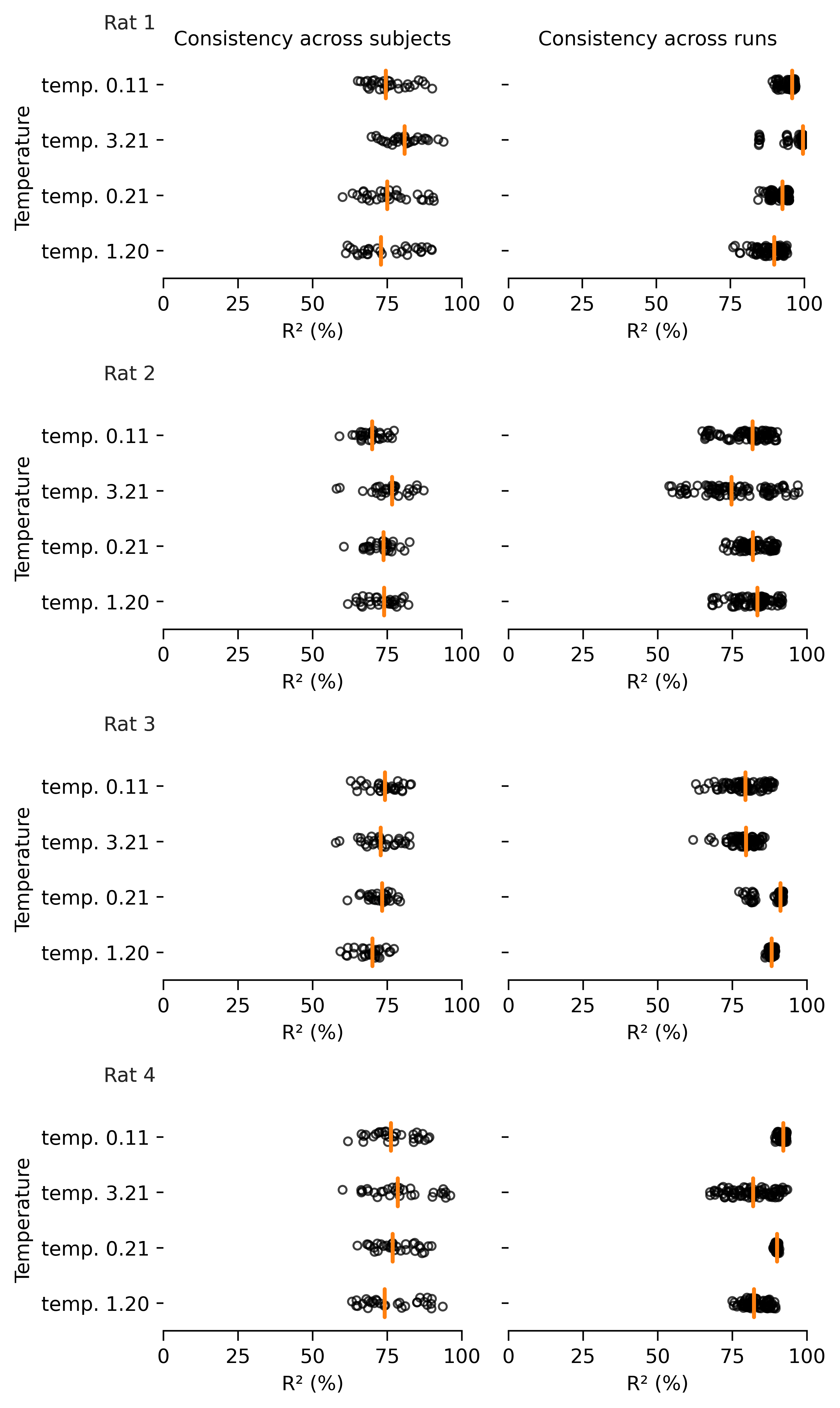
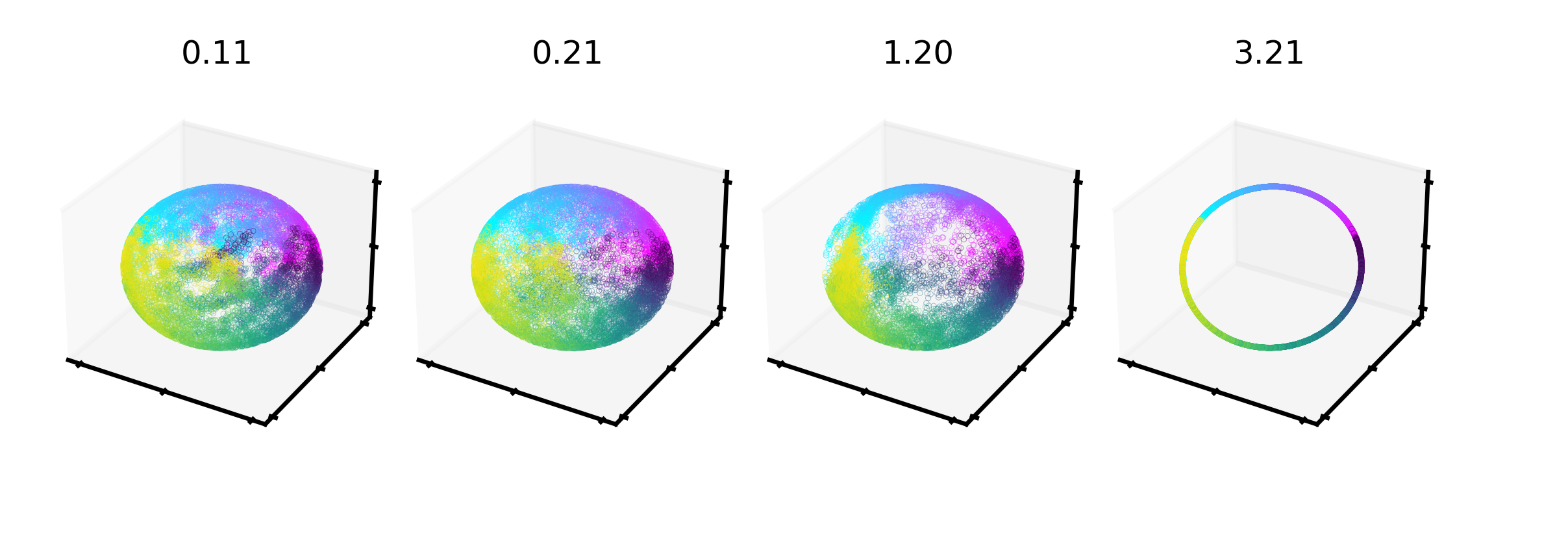
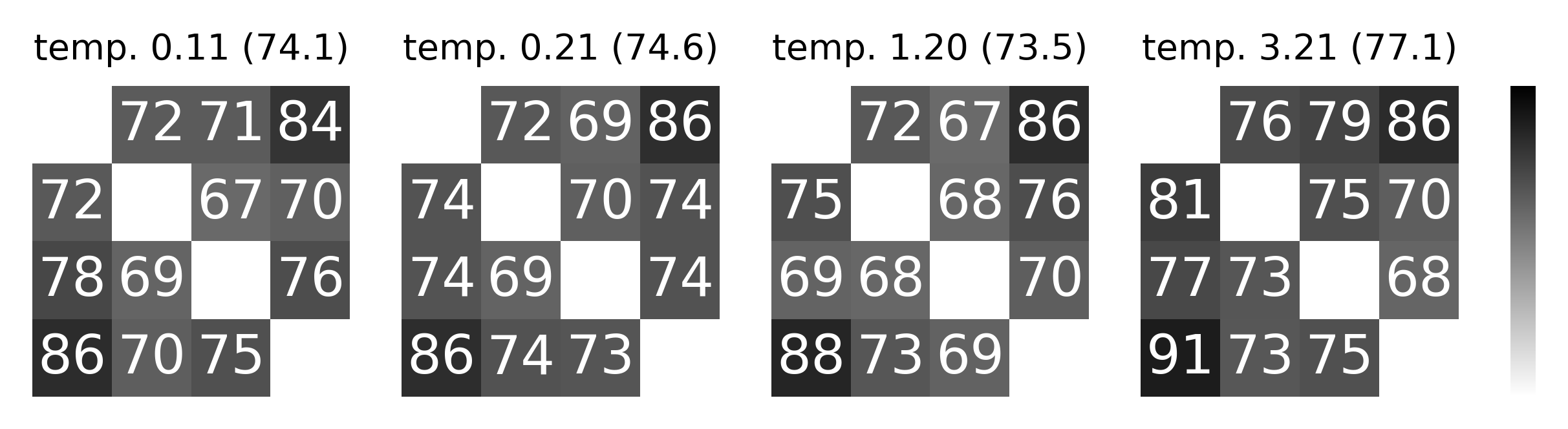
Extended Data Figure 2: Hyperparameter changes on visualization and consistency.#
Temperature has the largest effect on visualization (vs. consistency) of the embedding#
shown by a range from 0.1 to 3.21 (highest consistency for Rat 1), as can be appreciated in 3D (top) and post FastICA into a 2D embedding (middle). Bottom row shows the corresponding change on mean consistency.
Note, tempature is both a learnable and easily modified parameter in CEBRA.
[1]:
import pandas as pd
import numpy as np
from IPython.display import HTML
import matplotlib.pyplot as plt
def _rotate(angle):
rot = np.eye(2) * np.cos(angle)
rot[1, 0] = np.sin(angle)
rot[0, 1] = -rot[1, 0]
return rot
def scatter(data, index, ax, s=0.01, alpha=0.5, **kwargs):
mask = index[:, 1] > 0
ax.scatter(
*data[mask].T, c=index[mask, 0], s=s, cmap="viridis_r", alpha=alpha, **kwargs
)
ax.scatter(
*data[~mask].T, c=index[~mask, 0], s=s, cmap="cool", alpha=alpha, **kwargs
)
def plot_across_temperatures(embeddings):
display(HTML("<h2>3D CEBRA-Time</h2>"))
fig = plt.figure(figsize=(4, 4), dpi=600)
for i, args, embedding, data, ica, labels in embeddings.values:
data = data @ np.array([[0, 1, 0], [1, 0, 0], [0, 0, 1]])
ax = fig.add_subplot(1, 4, i + 1, projection="3d")
scatter(data, labels, ax)
ax.set_xlim([-1.1, 1.1])
ax.set_ylim([-1.1, 1.1])
ax.set_zlim([-1.1, 1.1])
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.set_zticklabels([])
ax.grid(visible=False)
ax.set_title("{temperature:.2f}".format(**args), fontsize=6)
fig.subplots_adjust(left=0, wspace=-0.0)
plt.show()
display(HTML("<h2>3D CEBRA-Time with FastICA -> 2D projection</h2>"))
fig = plt.figure(figsize=(4, 1), dpi=600)
for i, args, embedding, data, ica, labels in embeddings.values:
ax = fig.add_subplot(1, 4, i + 1)
rot = _rotate(np.pi)
scatter(ica @ rot, labels, ax, s=0.001, alpha=0.9, marker="o")
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.grid(visible=False)
ax.set_aspect("equal")
ax.axis("off")
ax.set_title("{temperature:.2f}".format(**args), fontsize=6)
plt.show()
embeddings = pd.read_hdf("../data/EDFigure2.h5", key="data")
plot_across_temperatures(embeddings)
3D CEBRA-Time
3D CEBRA-Time with FastICA -> 2D projection
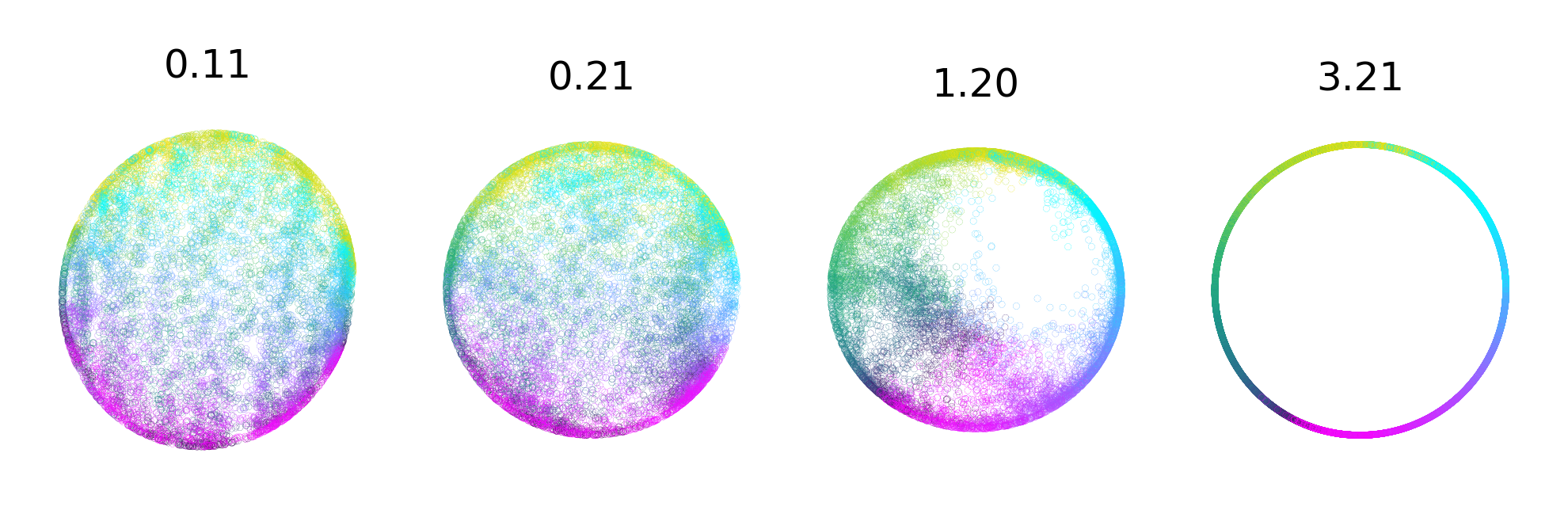
Orange line denotes the median and black dots are individual runs (subject consistency: 10 runs with 3 comparisons per rat; run consistency: 10 runs, each compared to 9 remaining runs).
[2]:
from matplotlib.markers import MarkerStyle
import warnings
import pathlib
import typing
import seaborn as sns
import matplotlib.pyplot as plt
ROOT = pathlib.Path("../data")
def recover_python_datatypes(element):
if isinstance(element, str):
if element.startswith("[") and element.endswith("]"):
if "," in element:
element = np.fromstring(element[1:-1], dtype=float, sep=",")
else:
element = np.fromstring(element[1:-1], dtype=float, sep=" ")
return element
def load_results(result_name):
"""Load a result file.
The first line in the result files specify the index columns,
the following lines are a CSV formatted file containing the
numerical results.
"""
results = {}
for result_csv in (ROOT / result_name).glob("*.csv"):
with open(result_csv) as fh:
index_names = fh.readline().strip().split(",")
df = pd.read_csv(fh).set_index(index_names)
df = df.applymap(recover_python_datatypes)
results[result_csv.stem] = df
return results
results_temperature = load_results("results_temperature")
[3]:
import seaborn as sns
import matplotlib.pyplot as plt
plt.rcParams["text.usetex"] = False
def to_cfm(values):
values = np.concatenate(values)
assert len(values) == 12, len(values)
c = np.zeros((4, 4))
c[:] = float("nan")
c[np.eye(4) == 0] = values
return c
fig, axs = plt.subplots(
ncols=5,
nrows=1,
figsize=(6, 1.25),
gridspec_kw={"width_ratios": [1, 1, 1, 1, 0.08]},
dpi=500,
)
last_ax = axs[-1]
for ax in axs:
ax.axis("off")
for ax, (key, log) in zip(axs[:-1], sorted(results_temperature.items())):
cfm = log.pivot_table(
"train", index=log.index.names, columns=["animal"], aggfunc="mean"
).apply(to_cfm, axis=1)
(cfm,) = cfm.values
sns.heatmap(
data=np.minimum(cfm * 100, 99),
vmin=20,
vmax=100,
xticklabels=[],
yticklabels=[],
cmap=sns.color_palette("gray_r", as_cmap=True),
annot=True,
annot_kws={"fontsize": 12},
cbar=True if (ax == axs[-2]) else False,
cbar_ax=last_ax if (ax == axs[-2]) else None,
ax=ax,
)
ax.set_title(f"{key} ({100*np.nanmean(cfm):.1f})", fontsize=8)
[4]:
from matplotlib.markers import MarkerStyle
import warnings
import typing
import itertools
def show_boxplot(df, metric, ax, labels=None):
sns.set_style("white")
with warnings.catch_warnings():
warnings.simplefilter("ignore")
color = "C1"
sns.boxplot(
data=df,
y="method",
x=metric,
orient="h",
order=labels, # unique(labels.values()),
# hue = "rat",
width=0.5,
color="k",
linewidth=2,
flierprops=dict(alpha=0.5, markersize=0, marker=".", linewidth=0),
medianprops=dict(
c="C1", markersize=0, marker=".", linewidth=2, solid_capstyle="round"
),
whiskerprops=dict(solid_capstyle="butt", linewidth=0),
# capprops = dict(c = 'C1', markersize = 0, marker = 'o', linewidth = 1),
showbox=False,
showcaps=False,
# shownotches = True
ax=ax,
)
marker_style = MarkerStyle("o", "none")
sns.stripplot(
data=df,
y="method",
x=metric,
orient="h",
size=4,
color="black",
order=labels,
marker=marker_style,
linewidth=1,
ax=ax,
alpha=0.75,
jitter=0.1,
zorder=-1,
)
ax.set_ylabel("")
sns.despine(left=True, bottom=False, ax=ax)
ax.tick_params(
axis="x", which="both", bottom=True, top=False, length=5, labelbottom=True
)
return ax
def _add_value(df, **kwargs):
for key, value in kwargs.items():
df[key] = value
return df
def join(results):
return pd.concat([_add_value(df, method=key) for key, df in results.items()])
metadata = [
("train", "Consistency across subjects", 100, "R² (%)", [0, 25, 50, 75, 100]),
(
"train_run_consistency",
"Consistency across runs",
100,
"R² (%)",
[0, 25, 50, 75, 100],
),
# Additional metrics, not plotted for space reasons.
# ("test_total_r2", "Decoding (direction, position)", 100, "R² (%)", [0,25,50,75,100]),
# ("test_position_error", "Decoding (positional error)", 100, "Error [cm]", [0, 10, 20])
]
results_ = join(results_temperature)
fig, axes = plt.subplots(
4, len(metadata), figsize=(3 * len(metadata), 10), dpi=500, sharey=True
)
label_order = tuple(results_temperature.keys())
def _agg(v):
return sum(v) / len(v)
for metric_id, (metric, metric_name, scale, xlabel, xlim) in enumerate(metadata):
table = (
results_.reset_index(drop=True)
.pivot_table(
metric,
index=["animal", "repeat"],
columns=["method"],
aggfunc=list, # lambda v : list(itertools.chain.from_iterable(v) if isinstance(v, list) else list(v))
)
.applymap(
lambda v: list(
itertools.chain.from_iterable(v)
if isinstance(v[0], typing.Iterable)
else v
)
)
.groupby("animal", level=0)
.agg(lambda v: np.stack(v).mean(0))
)
for animal in table.index:
df = table.loc[animal].reset_index()
df.columns = "method", "metric"
df = df.explode("metric")
df["metric"] *= scale
show_boxplot(
df=df, metric="metric", ax=axes[animal, metric_id], labels=label_order
)
ax = axes[animal, metric_id]
ax.set_xlabel(xlabel)
ax.set_xticks(xlim)
ax.spines["bottom"].set_bounds(min(xlim), max(xlim))
axes[0, metric_id].set_title(metric_name, fontsize=10)
axes[animal, 0].set_ylabel(f"Temperature")
axes[animal, 0].text(-20, -1, f"Rat {animal+1}")
plt.tight_layout()
plt.show()