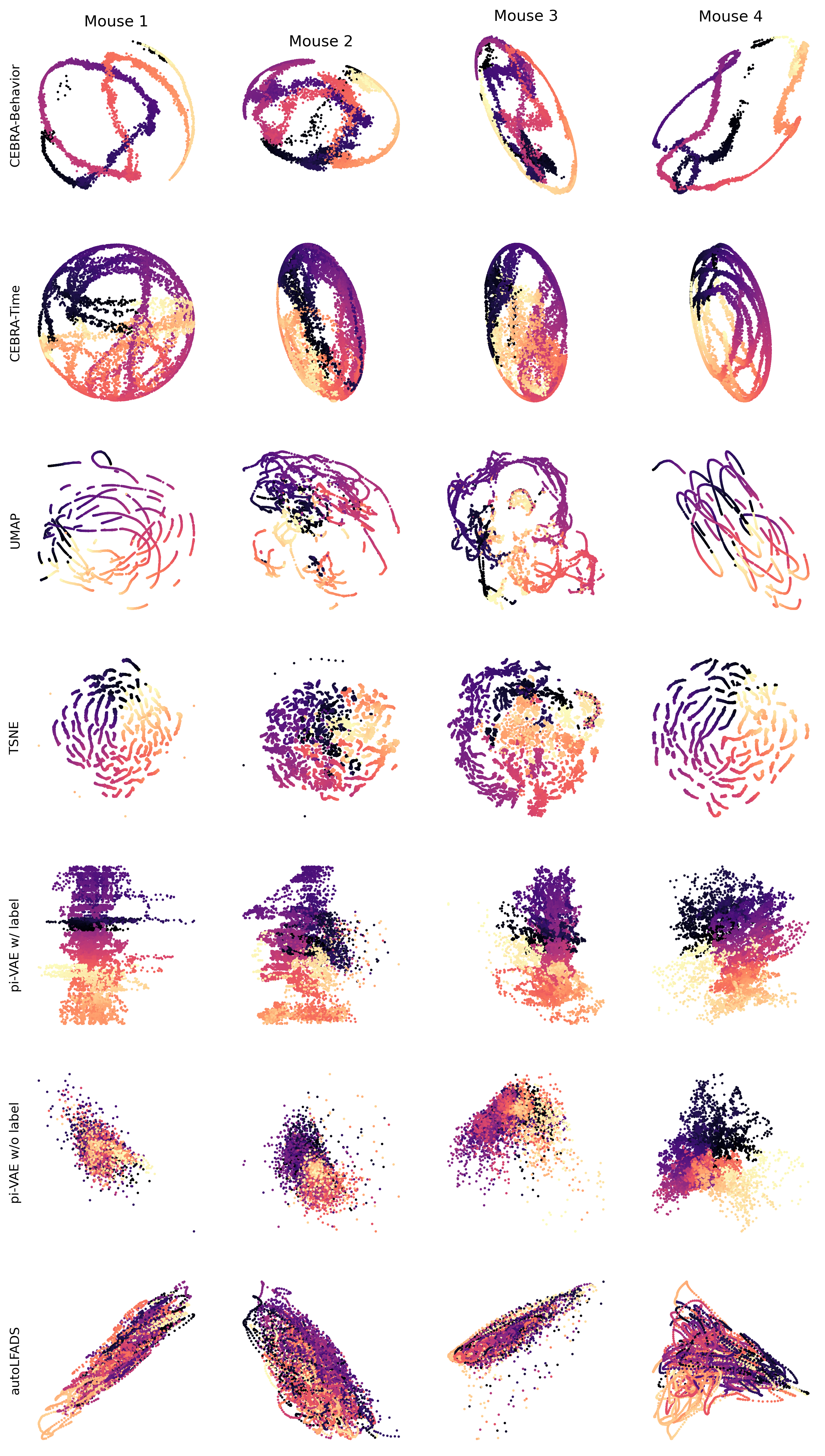
Extended Data Figure 9: CEBRA produces consistent, highly decodable embeddings#
import plot and data loading dependencies#
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sklearn.linear_model
[2]:
data = pd.read_hdf("../data/EDFigure9.h5", key="data")
Additional 4 sessions with the most neurons in the Allen visual dataset calcium recording shown for all algorithms we benchmarked#
For CEBRA-Behavior and \cebra-Time, we used temperature 1, time offset 10, batch size 128 and 10k training steps. For UMAP, we used cosine metric and \(n\_neighbors\) 15 and \(min\_dist\) 0.1. For tSNE, we used cosine metric and \(perplexity\) 30. For conv-pi-VAE, we trained with 600 epochs, batch size 200 and learning rate \(5\times 10^{-4}\). All methods used 10 time bins input. CEBRA was trained with 3D latent and all other methods were obtained with 2D latent dimension.
[3]:
embedding_idx = list(range(7))
fig = plt.figure(figsize=(11, 20 / 7 * len(embedding_idx)), dpi = 300)
emissions_list = [
data["cebra"],
data["emission_cebra_time"],
data["emission_umap"],
data["emission_tsne"],
data["emission_pivae_w"],
data["emission_pivae_wo"],
data["emission_autolfads"],
]
for i in range(4):
for j_, j in enumerate(embedding_idx):
ax = fig.add_subplot(len(embedding_idx), 4, j_ * 4 + i + 1)
idx1, idx2 = (0, 1)
if j <= 4:
lin = sklearn.linear_model.LinearRegression()
lin.fit(emissions_list[j][i], emissions_list[j][0])
fitted = lin.predict(emissions_list[j][i])
else:
lin = sklearn.linear_model.LinearRegression()
lin.fit(emissions_list[j][i], emissions_list[j][3])
fitted = lin.predict(emissions_list[j][i])
ax.scatter(
fitted[:, idx1],
fitted[:, idx2],
c=np.tile(np.arange(900), 10)[:len(fitted)],
cmap="magma",
s=.75,
)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["bottom"].set_visible(False)
ax.spines["left"].set_visible(False)
ax.set_xticks([])
ax.set_yticks([])
if j <= 1:
ax.set_aspect("equal")
if j == 0:
plt.title(["Mouse 1", "Mouse 2", "Mouse 3", "Mouse 4"][i])
if i == 0:
plt.ylabel(
[
"CEBRA-Behavior",
"CEBRA-Time",
"UMAP",
"TSNE",
"pi-VAE w/ label",
"pi-VAE w/o label",
"autoLFADS"
][j],
)
#plt.subplots_adjust(wspace = .1)
#plt.savefig('fig.png', bbox_inches = 'tight', transparent = True)