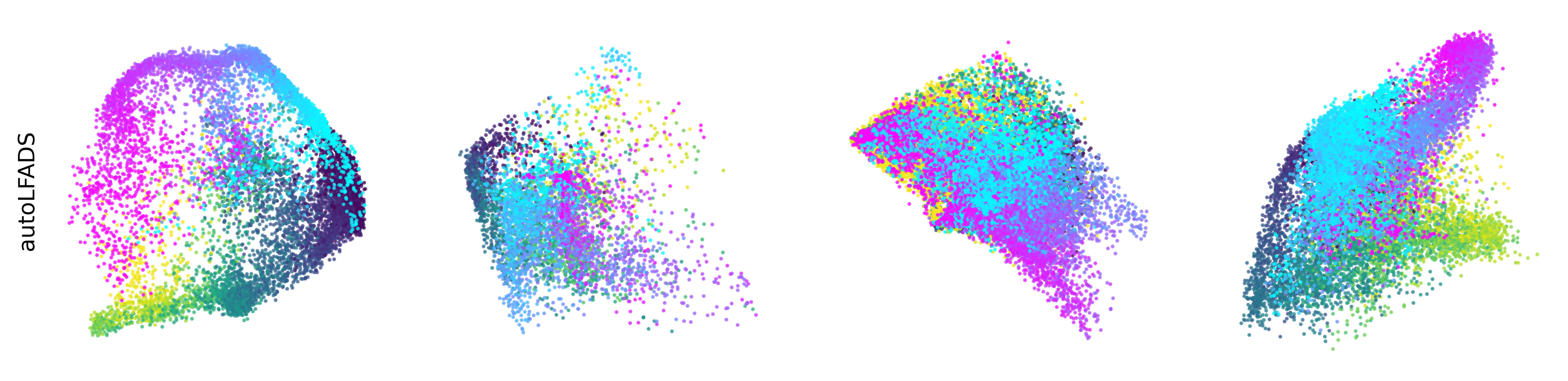
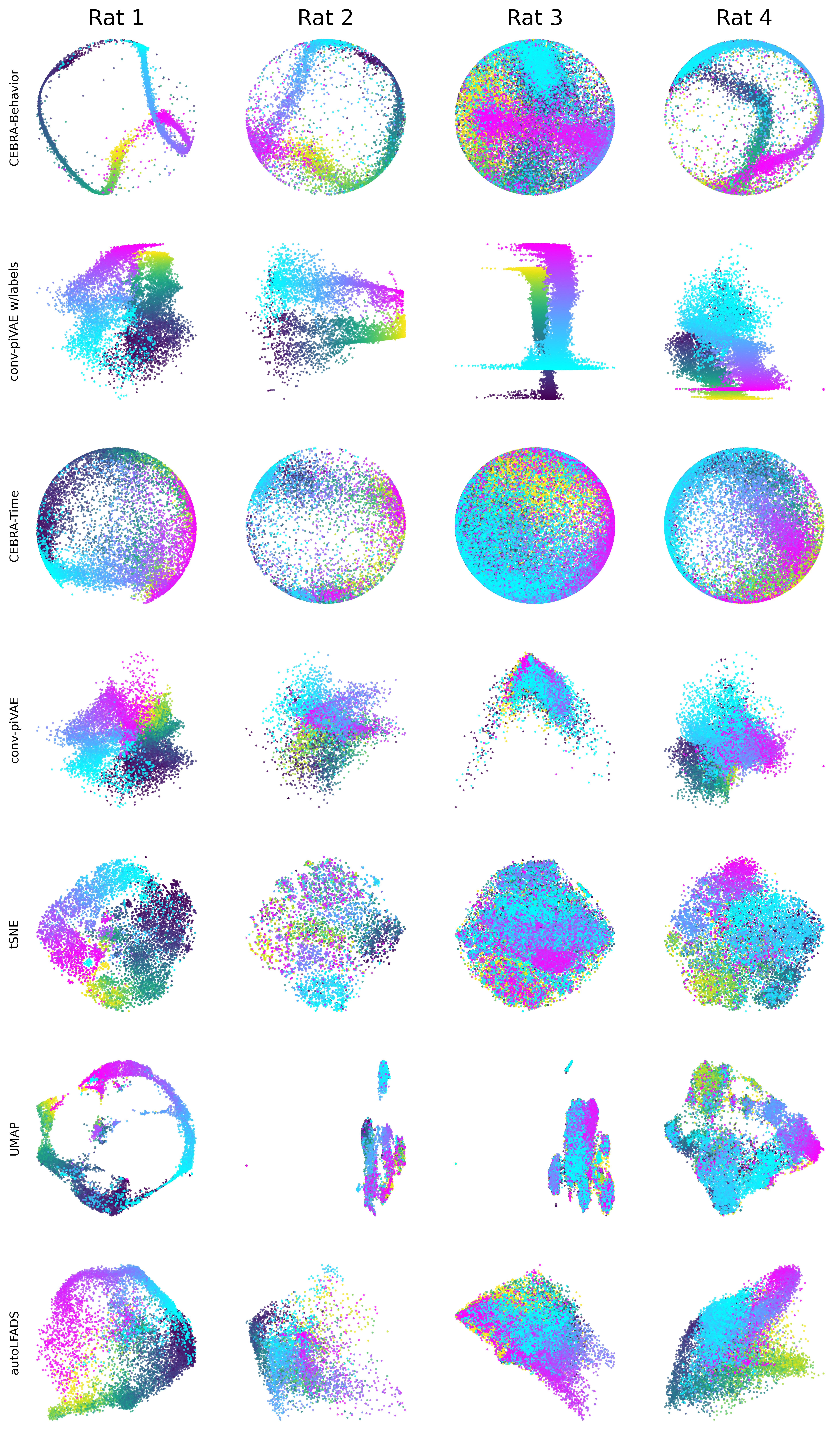
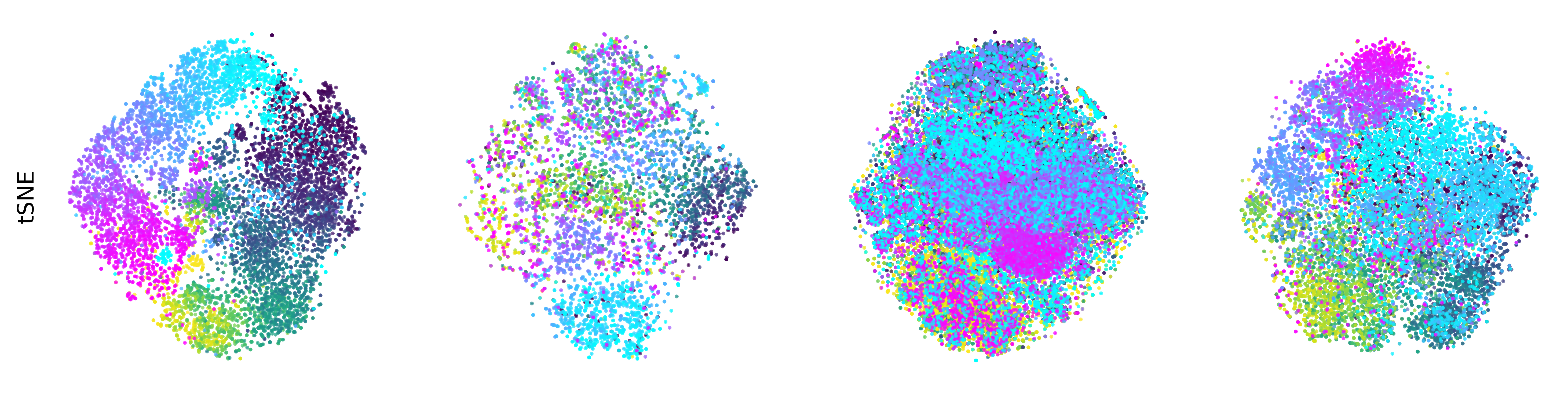
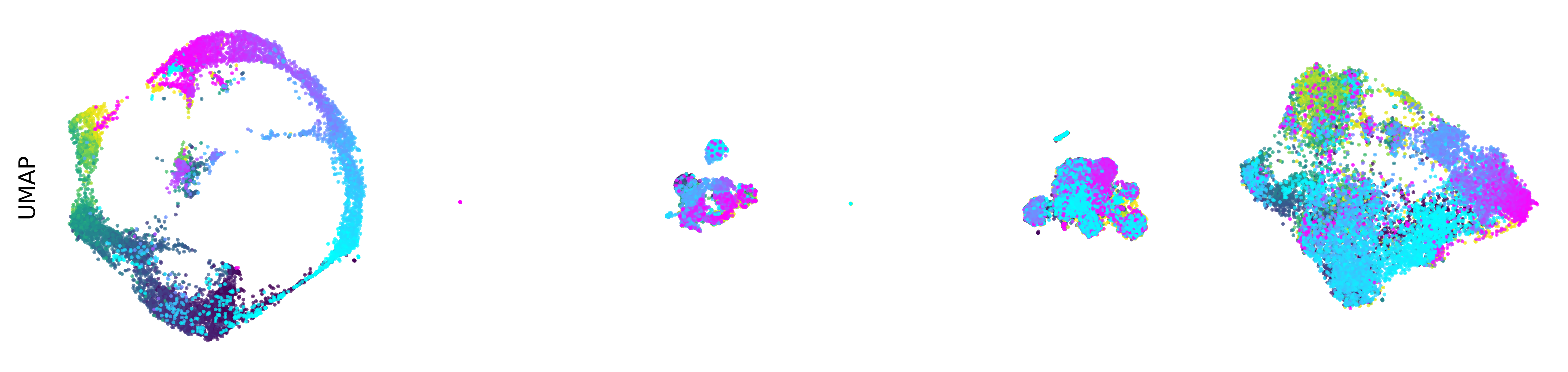
Extended Data Figure 3: CEBRA produces consistent, highly decodable embeddings#
Additional rat data shown for all algorithms we benchmarked (see Methods). CEBRA was trained with output latent on the 2-sphere (the minimum) and all other methods were obtained with a 2D latent in Euclidean space.
[1]:
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.concat([
pd.read_hdf("../data/EDFigure3.h5", key="data"),
pd.read_hdf("../data/EDFigure3_addition.h5", key="data")
], axis = 0, ignore_index = True)
def scatter(data, index, ax, s=0.01, alpha=0.5):
mask = index[:, 1] > 0
ax.scatter(*data[mask].T, c=index[mask, 0], s=s, cmap="viridis", alpha=alpha)
ax.scatter(*data[~mask].T, c=index[~mask, 0], s=s, cmap="cool", alpha=alpha)
fig = plt.figure(figsize=(4 * 3, 7 * 3), dpi=600)
for i in df.index:
ax = fig.add_subplot(7, 4, i + 1)
scatter(df.loc[i, "emission"][:, :2], df.loc[i, "labels"], ax=ax, s=0.5, alpha=0.7)
ax.set_yticklabels([])
ax.set_xticklabels([])
ax.set_xticks([])
ax.set_yticks([])
sns.despine(bottom=True, left=True, ax=ax)
# first row labels
if i // 4 == 0:
ax.set_title(f"Rat {df.loc[i, 'animal']}", fontsize=18)
# first column labels
if i % 4 == 0:
ax.set_ylabel(df.loc[i, "method"])
For a higher resolution plot, we export each row as a separate file:
[2]:
def scatter(data, index, ax, s=0.01, alpha=0.5):
mask = index[:, 1] > 0
ax.scatter(*data[mask].T, c=index[mask, 0], s=s, cmap="viridis", alpha=alpha)
ax.scatter(*data[~mask].T, c=index[~mask, 0], s=s, cmap="cool", alpha=alpha)
def export_highres():
for method in df.method.unique():
print(method)
fig = plt.figure(figsize=(4 * 3, 1 * 3), dpi=600)
entry = df[df.method == method].set_index("animal")
for i, animal in enumerate(sorted(entry.index)):
ax = fig.add_subplot(1, 4, i + 1)
scatter(
entry.loc[animal, "emission"][:, :2],
entry.loc[animal, "labels"],
ax=ax, s=0.5, alpha=0.7
)
ax.set_yticklabels([])
ax.set_xticklabels([])
ax.set_xticks([])
ax.set_yticks([])
ax.set_aspect("equal")
sns.despine(bottom=True, left=True, ax=ax)
# first row labels
#if i // 4 == 0:
# ax.set_title(f"Rat {df.loc[i, 'animal']}")
# first column labels
if i % 4 == 0:
ax.set_ylabel(method)
method = method.replace('/', '-')
plt.savefig(f'edf3_{method}.png', bbox_inches = "tight", transparent = True)
plt.show()
export_highres()
CEBRA-Behavior
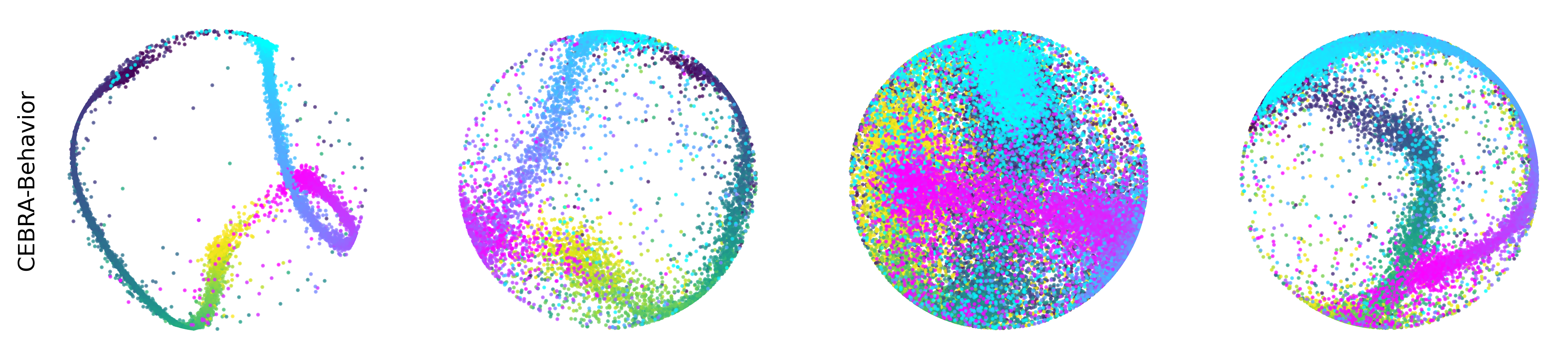
conv-piVAE w/labels
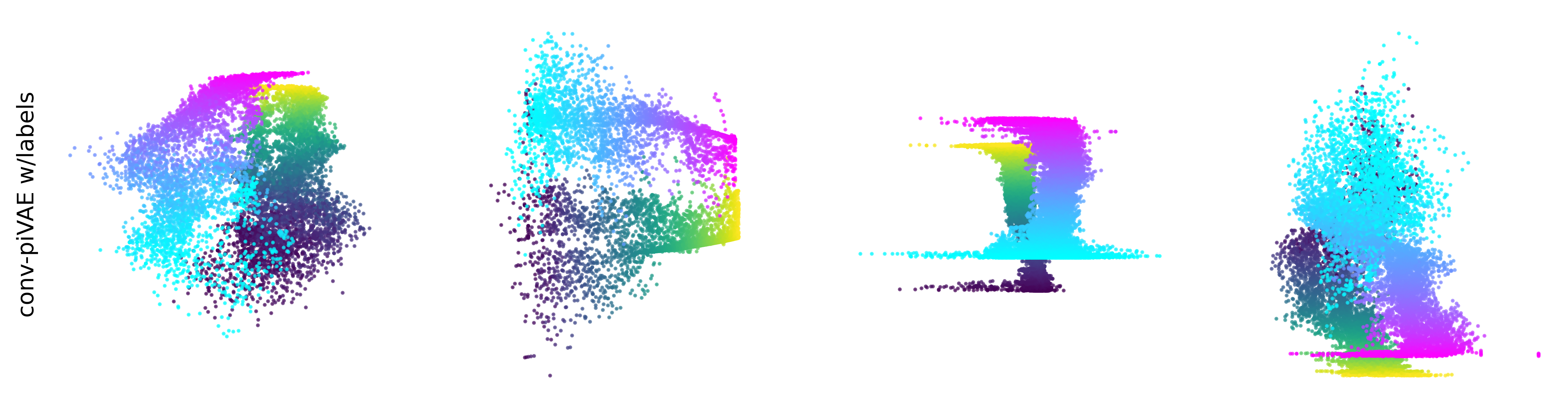
CEBRA-Time
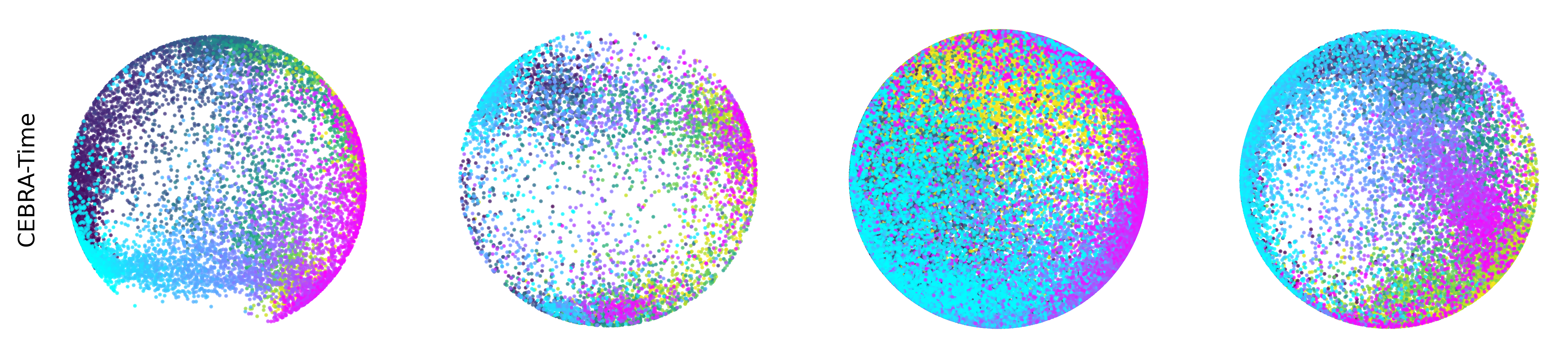
conv-piVAE
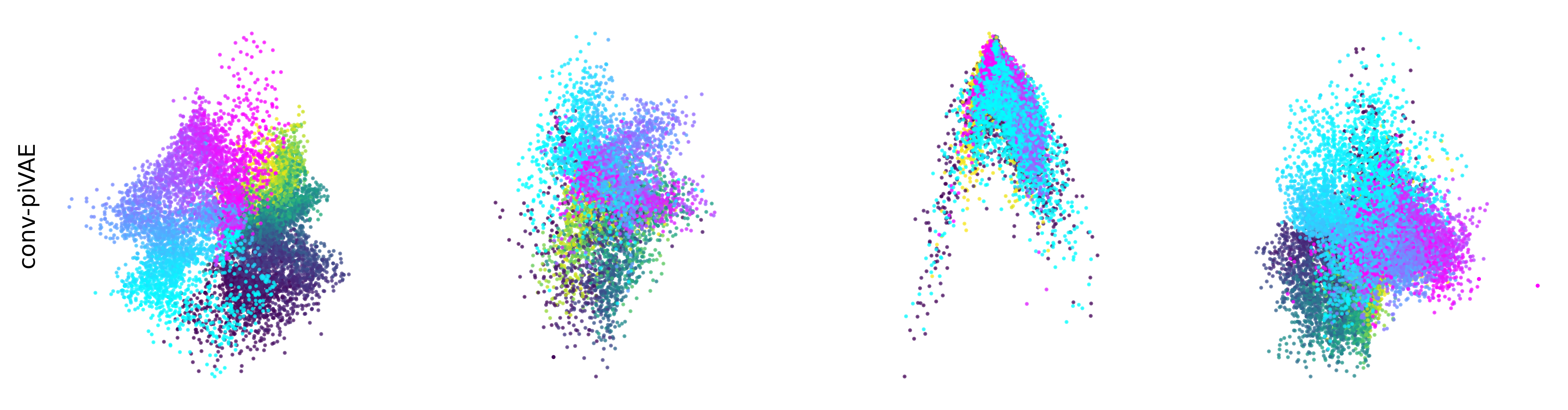
tSNE
UMAP
autoLFADS