Extended Data Figure 5: Hypothesis testing with CEBRA.#
Example data from a hippocampus recording session (Rat 1).#
We test possible relationships between three experimental variables (rat location, velocity, movement direction) and the neural recordings (120 neurons, not shown).
[1]:
import glob
import sys
import numpy as np
import itertools
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import torch
import json
def load_emission(path, which):
emission = torch.load(path)
return emission[which]
def load_loss(path):
emission = torch.load(path, map_location="cpu")
return emission["loss"]
def load_args(path):
with open(path, "r") as fh:
idx = [5, 7, 9, -1]
args = json.load(fh)
position, velocity, direction, loader = [
args["data"].split("-")[i] for i in idx
]
args["direction"] = direction
args["velocity"] = velocity
args["position"] = position
args["loader"] = loader
return args, (position, velocity, direction, loader, args["num_output"])
def prepare_data():
keys = ["position", "velocity", "direction"]
options = list(itertools.product(keys, ["original", "shuffle"]))
options = (
np.array(options).reshape(3, 2, 2).transpose(1, 0, 2).reshape(6, 2).tolist()
)
options = list(map(tuple, options))
print(options)
import pandas as pd
def rep(k):
if len(k) == 0:
return "none"
if len(k) == 2:
return "none"
return k.pop()
args = []
for selection in itertools.product(options, options):
kwargs = {opt: set() for opt in keys}
for k, v in selection:
kwargs[k].add(v)
args.append(
{
"src": tuple(selection[0]),
"tgt": tuple(selection[1]),
"config": tuple(rep(kwargs[k]) for k in keys),
}
)
df = pd.DataFrame(args).pivot("src", "tgt")
return df
def scatter(data, ax):
pca = PCA(2, whiten=False)
pca.fit(data)
data = pca.transform(data)[:, :]
data /= data.max(keepdims=True)
x, y = data[:, :2].T
ax.scatter(x, y, s=1, alpha=0.25, edgecolor="none", c="k") # , c = f"C{n}")
def apply_style(ax):
sns.despine(ax=ax, trim=True, left=True, bottom=True)
ax.set_xlim([-1, 1])
ax.set_ylim([-1, 1])
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.tick_params(axis="both", which="both", length=0)
ax.set_aspect("equal")
def scatter_grid(df, dim):
fig, axes = plt.subplots(6, 6, figsize=(5, 5), dpi=200)
df = df.iloc[[0, 2, 4, 1, 3, 5], [0, 2, 4, 1, 3, 5]]
for i, (index, row) in enumerate(zip(df.index, axes)):
for j, (column, ax) in enumerate(zip(df.columns, row)):
apply_style(ax)
if i == 5:
ax.set_xlabel("{}\n{}".format(*column[1]), fontsize=6)
if j == 0:
ax.set_ylabel("{}\n{}".format(*index), fontsize=6)
if j > i:
continue
entry = df[column][index]
key = entry + ("continuous", dim, "train")
if entry == ("none", "none", "none"):
continue
emission = files[key]
scatter(emission[:, :], ax=ax)
def plot_heatmap(df, dim):
def to_heatmap(entry):
key = entry + ("continuous", dim, "loss")
return min(files.get(key, [float("nan")]))
df = df.iloc[[0, 2, 4, 1, 3, 5], [0, 2, 4, 1, 3, 5]]
labels = ["{}\n{}".format(*i) for i in df.index]
hm = df.applymap(to_heatmap)
for i, j in itertools.product(range(len(hm)), range(len(hm))):
if j > i:
hm.iloc[i, j] = float("nan")
plt.figure(figsize=(4, 4), dpi=150)
sns.heatmap(
hm,
cmap="gray",
xticklabels=labels,
yticklabels=labels,
annot=True,
fmt=".2f",
square=True,
)
plt.xlabel("")
plt.ylabel("")
plt.gca().set_xticklabels(labels, fontsize=8)
plt.gca().set_yticklabels(labels, fontsize=8)
plt.show()
df = prepare_data()
emission_files = sorted(glob.glob("../data/EDFigure5/*/emission*"))
arg_files = sorted(glob.glob("../data/EDFigure5/*/args.json"))
losses = [load_emission(e, "losses") for e in emission_files]
emissions = [load_emission(e, "train") for e in emission_files]
emissions_test = [load_emission(e, "test") for e in emission_files]
args = [load_args(path) for path in arg_files]
files = {keys + ("train",): values for (_, keys), values in zip(args, emissions)}
files.update(
{keys + ("test",): values for (_, keys), values in zip(args, emissions_test)}
)
files.update({keys + ("loss",): values for (_, keys), values in zip(args, losses)})
[('position', 'original'), ('velocity', 'original'), ('direction', 'original'), ('position', 'shuffle'), ('velocity', 'shuffle'), ('direction', 'shuffle')]
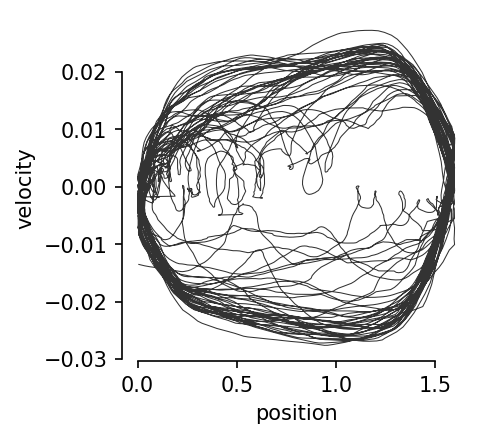
Relationship between velocity and position.#
[2]:
import os
import joblib
def prepare_dataset(cache="../data/EDFigure5/achilles_behavior.jl"):
"""If you have CEBRA installed, you can adapt this function to load other
rats, or adapt the data loading in other ways. Otherwise, pre-computed data
will be loaded."""
if not os.path.exists(cache):
import cebra.datasets
DATADIR = "/home/stes/projects/neural_cl/cebra_public/data"
dataset = cebra.datasets.init(
"rat-hippocampus-achilles-3fold-trial-split-0", root=DATADIR
)
index = dataset.index
joblib.dump(index, cache)
return joblib.load(cache)
[3]:
import scipy.signal
import matplotlib.pyplot as plt
import seaborn as sns
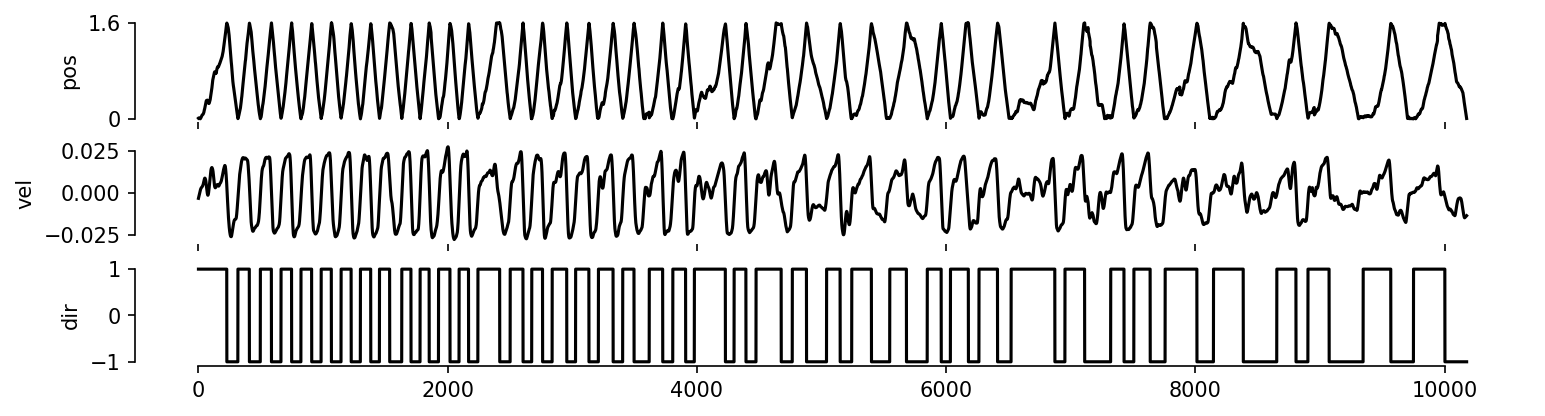
def dataset_plots(index):
"""Make a summary plot of the hippocampus dataset.
Args:
index is the [position, left, right] behavior index also used for training
CEBRA models.
"""
# preprocess the dataset
position = index[:, 0]
velocity = scipy.signal.savgol_filter(
index[:, 0], window_length=31, polyorder=2, deriv=1
)
accel = scipy.signal.savgol_filter(
index[:, 0], window_length=11, polyorder=2, deriv=2
)
direction = index[:, 1] - index[:, 2]
# plot figure
fig, axes = plt.subplots(3, 1, figsize=(12, 3), sharex=True, dpi=150)
axes[0].plot(index[:, 0], c="k")
axes[0].set_ylim([-0.05, 1.65])
axes[0].set_yticks([0, 1.6])
axes[0].set_yticklabels([0, 1.6])
axes[1].plot(velocity, c="k")
axes[2].plot(direction, c="k")
for i, ax in enumerate(axes):
sns.despine(ax=ax, trim=True, bottom=i != 2)
for ax, lbl in zip(axes, ["pos", "vel", "dir"]):
ax.set_ylabel(lbl)
plt.show()
plt.figure(figsize=(3, 3), dpi=150)
plt.plot(index[:, 0], velocity, linewidth=0.5, color="black", alpha=0.8)
plt.xlabel("position")
plt.ylabel("velocity")
sns.despine(trim=True)
plt.show()
index = prepare_dataset()
dataset_plots(index)
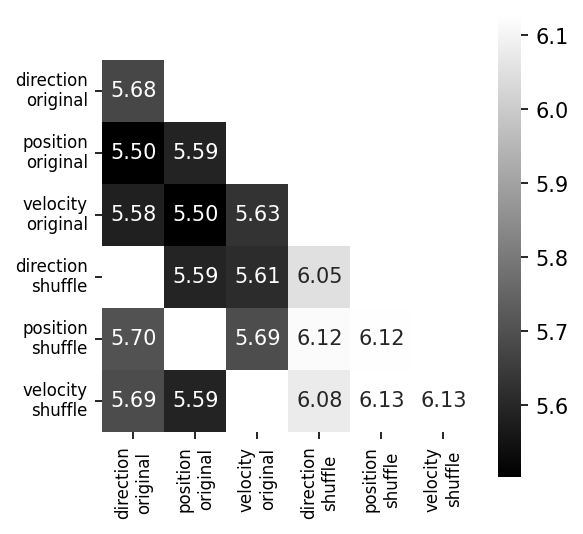
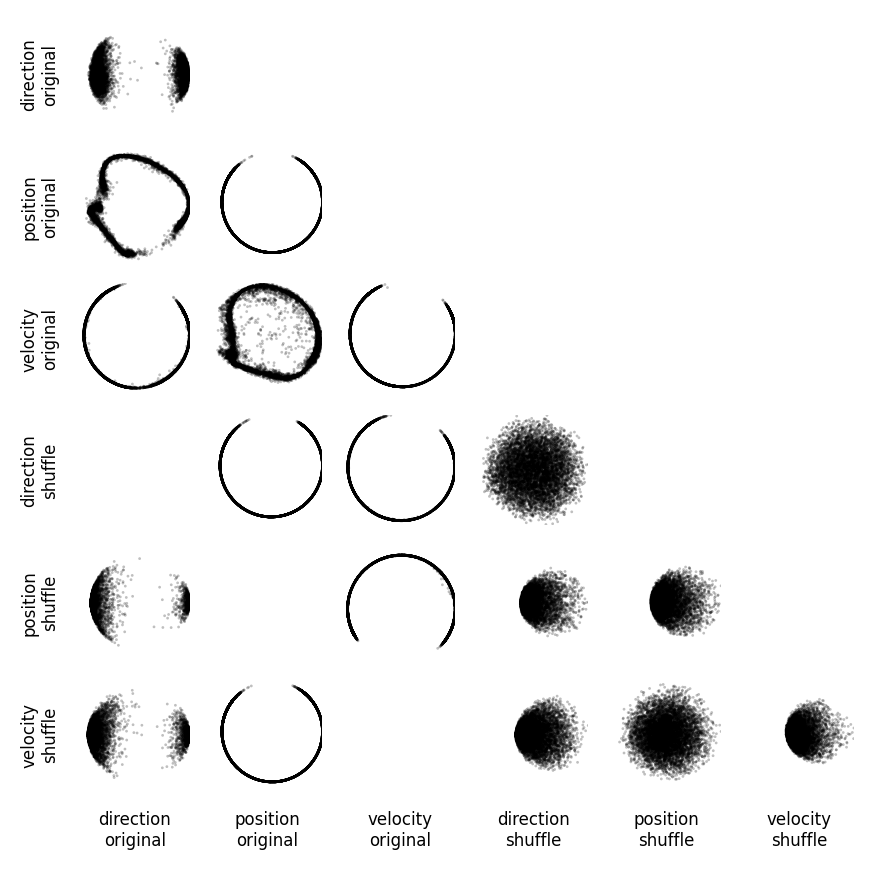
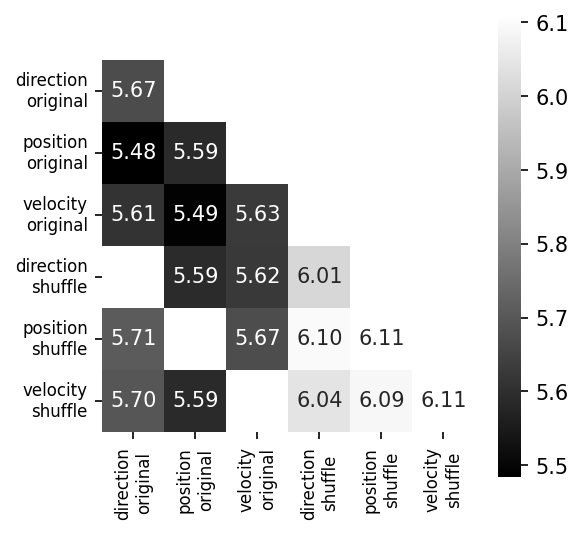
Training CEBRA with three-dimensional outputs on every single experimental variable (main diagonal) and every combination of two variables.#
All variables are treated as “continuous” in this experiment. We compare original to shuffled variables (shuffling is done by permuting all samples over the time dimension) as a control. We project the original three dimensional space onto the first principal components. We show the minimum value of the InfoNCE loss on the trained embedding for all combinations in the confusion matrix (lower number is better). Either velocity or direction, paired with position information is needed for maximum structure in the embedding (highlighted, colored), yielding lowest InfoNCE error.
[4]:
scatter_grid(df, 3)
plot_heatmap(df, 3)

Using an eight-dimensional CEBRA embedding does not qualitatively alter the results.#
We again report the first two principal components as well as InfoNCE training error upon convergence, and find non-trivial embeddings with lowest training error for combinations of direction/velocity and position.
[5]:
scatter_grid(df, 8)
plot_heatmap(df, 8)
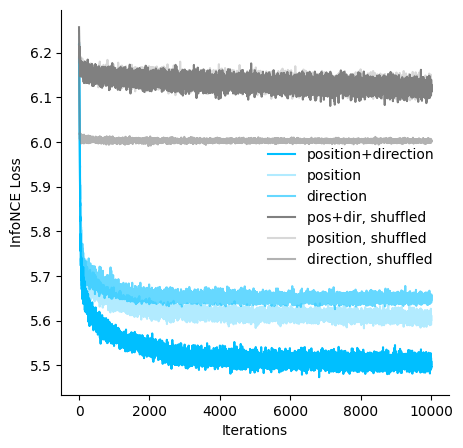
[6]:
hypothesis_loss = joblib.load("../data/EDFigure5/hypothesis_testing_loss.jl")
fig = plt.figure(figsize=(5, 5))
ax = plt.subplot(111)
ax.plot(hypothesis_loss["posdir"], c="deepskyblue", label="position+direction")
ax.plot(hypothesis_loss["pos"], c="deepskyblue", alpha=0.3, label="position")
ax.plot(hypothesis_loss["dir"], c="deepskyblue", alpha=0.6, label="direction")
ax.plot(hypothesis_loss["posdir-shuffled"], c="gray", label="pos+dir, shuffled")
ax.plot(
hypothesis_loss["pos-shuffled"], c="gray", alpha=0.3, label="position, shuffled"
)
ax.plot(
hypothesis_loss["dir-shuffled"], c="gray", alpha=0.6, label="direction, shuffled"
)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.set_xlabel("Iterations")
ax.set_ylabel("InfoNCE Loss")
plt.legend(bbox_to_anchor=(0.5, 0.3), frameon=False)
plt.show()
[7]:
hypothesis_decode = joblib.load("../data/EDFigure5/hypothesis_testing_decode.jl")
[8]:
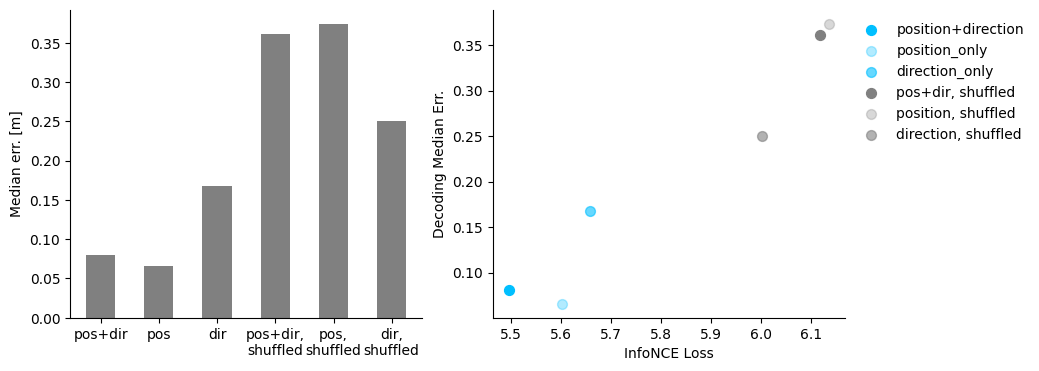
fig = plt.figure(figsize=(10, 4))
ax1 = plt.subplot(121)
ax1.bar(
np.arange(6),
[
hypothesis_decode["posdir"],
hypothesis_decode["pos"],
hypothesis_decode["dir"],
hypothesis_decode["posdir-shuffled"],
hypothesis_decode["pos-shuffled"],
hypothesis_decode["dir-shuffled"],
],
width=0.5,
color="gray",
)
ax1.spines["top"].set_visible(False)
ax1.spines["right"].set_visible(False)
ax1.set_xticks(np.arange(6))
ax1.set_xticklabels(
["pos+dir", "pos", "dir", "pos+dir,\nshuffled", "pos,\nshuffled", "dir,\nshuffled"]
)
ax1.set_ylabel("Median err. [m]")
ax2 = plt.subplot(122)
ax2.scatter(
hypothesis_loss["posdir"][-1],
hypothesis_decode["posdir"],
s=50,
c="deepskyblue",
label="position+direction",
)
ax2.scatter(
hypothesis_loss["pos"][-1],
hypothesis_decode["pos"],
s=50,
c="deepskyblue",
alpha=0.3,
label="position_only",
)
ax2.scatter(
hypothesis_loss["dir"][-1],
hypothesis_decode["dir"],
s=50,
c="deepskyblue",
alpha=0.6,
label="direction_only",
)
ax2.scatter(
hypothesis_loss["posdir-shuffled"][-1],
hypothesis_decode["posdir-shuffled"],
s=50,
c="gray",
label="pos+dir, shuffled",
)
ax2.scatter(
hypothesis_loss["pos-shuffled"][-1],
hypothesis_decode["pos-shuffled"],
s=50,
c="gray",
alpha=0.3,
label="position, shuffled",
)
ax2.scatter(
hypothesis_loss["dir-shuffled"][-1],
hypothesis_decode["dir-shuffled"],
s=50,
c="gray",
alpha=0.6,
label="direction, shuffled",
)
ax2.spines["top"].set_visible(False)
ax2.spines["right"].set_visible(False)
ax2.set_xlabel("InfoNCE Loss")
ax2.set_ylabel("Decoding Median Err.")
plt.legend(bbox_to_anchor=(1, 1), frameon=False)
plt.show()