Figure 2: Hypothesis-driven and discovery-driven analysis with CEBRA#
For reference, here is the full figure

import plot and data loading dependencies#
[1]:
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
import pandas as pd
import numpy as np
import pathlib
[2]:
data = pd.read_hdf("../data/Figure2.h5", key="data")
data_fig_2d = pd.read_hdf("../data/SupplVideo1.h5", key="data")
Figure 2b#
CEBRA with position-hypothesis derived embedding, shuffled (erroneous), time-only, and Time+Behavior (hybrid; here, a 5D space was used, where first 3D is guided by both behavior+time, and last 2D is guided only by time, and the first 3 latents are plotted).
[3]:
method_viz = data["visualization"]
fig = plt.figure(figsize=(20, 5))
for i, model in enumerate(["hypothesis", "shuffled", "discovery", "hybrid"]):
ax = fig.add_subplot(1, 4, i + 1, projection="3d")
emb = method_viz[model]["embedding"]
label = method_viz[model]["label"]
r = label[:, 1] == 1
l = label[:, 2] == 1
idx1, idx2, idx3 = (0, 1, 2)
if i == 3:
idx1, idx2, idx3 = (1, 2, 0)
ax.scatter(
emb[l, idx1], emb[l, idx2], emb[l, idx3], c=label[l, 0], cmap="cool", s=0.1
)
ax.scatter(emb[r, idx1], emb[r, idx2], emb[r, idx3], c=label[r, 0], s=0.1)
ax.axis("off")
ax.set_title(f"{model}", fontsize=20)
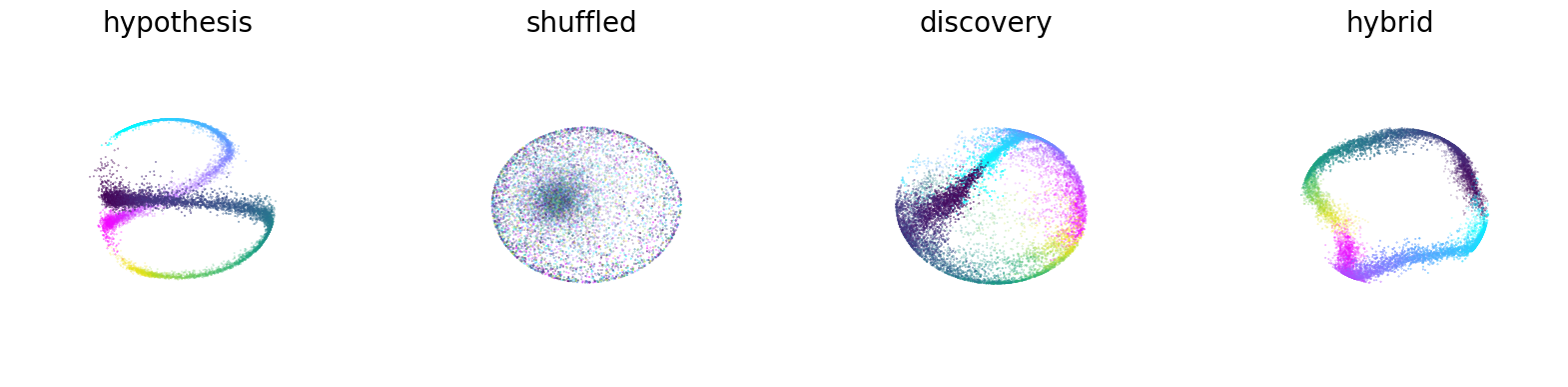
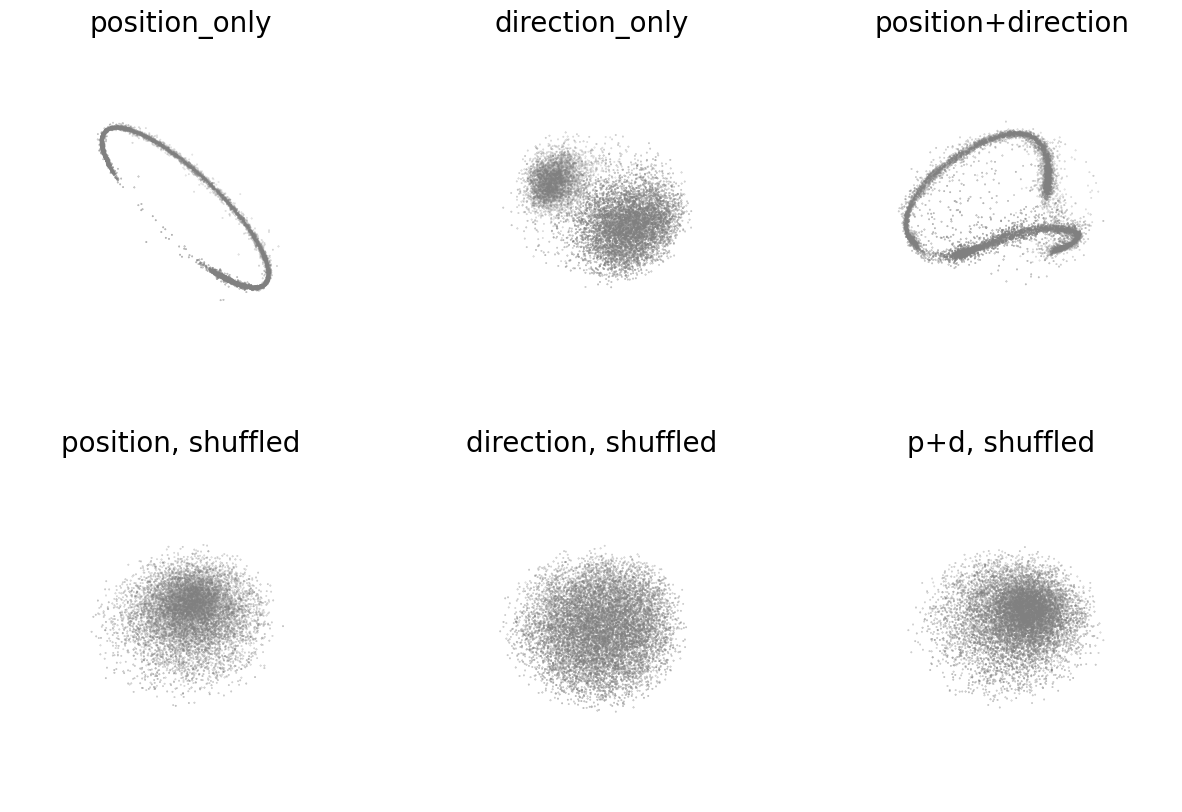
Figure 2c#
Embeddings with position-only, direction-only, and shuffled position-only, direction-only for hypothesis testing. The loss function can be used as a metric for embedding quality.
[4]:
hypothesis_viz = data["hypothesis_testing"]["viz"]
fig = plt.figure(figsize=(15, 10))
titles = {
"pos": "position_only",
"dir": "direction_only",
"posdir": "position+direction",
"pos-shuffled": "position, shuffled",
"dir-shuffled": "direction, shuffled",
"posdir-shuffled": "p+d, shuffled",
}
for i, model in enumerate(
["pos", "dir", "posdir", "pos-shuffled", "dir-shuffled", "posdir-shuffled"]
):
emb = hypothesis_viz[model]
ax = fig.add_subplot(2, 3, i + 1, projection="3d")
idx1, idx2, idx3 = (0, 1, 2)
ax.scatter(emb[:, idx1], emb[:, idx2], emb[:, idx3], c="gray", s=0.1)
ax.axis("off")
ax.set_title(f"{titles[model]}", fontsize=20)
[5]:
hypothesis_loss = data["hypothesis_testing"]["loss"]
fig = plt.figure(figsize=(4, 4))
ax = plt.subplot(111)
titles = {
"pos": "position_only",
"dir": "direction_only",
"posdir": "position+direction",
"pos-shuffled": "position, shuffled",
"dir-shuffled": "direction, shuffled",
"posdir-shuffled": "p+d, shuffled",
}
alphas = {
"pos": 0.3,
"dir": 0.6,
"posdir": 1,
"pos-shuffled": 0.3,
"dir-shuffled": 0.6,
"posdir-shuffled": 1,
}
for model in [
"pos",
"dir",
"posdir",
"pos-shuffled",
"dir-shuffled",
"posdir-shuffled",
]:
if "shuffled" in model:
c = "gray"
else:
c = "deepskyblue"
ax.plot(hypothesis_loss[model], c=c, alpha=alphas[model], label=titles[model])
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.set_xlabel("Iterations", fontsize=15)
ax.set_ylabel("InfoNCE Loss", fontsize=15)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.legend(bbox_to_anchor=(1, 0.5), frameon=False, fontsize=10)
[5]:
<matplotlib.legend.Legend at 0x7fe2fd099cf0>
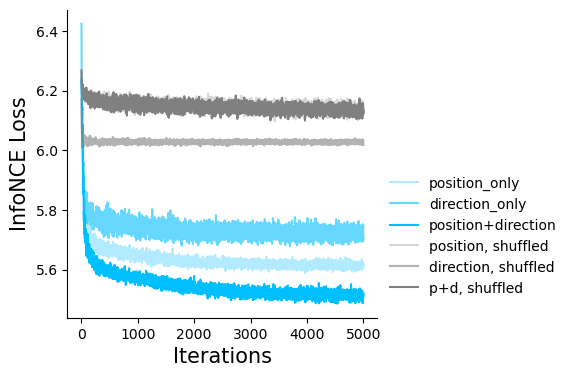
Figure 2d#
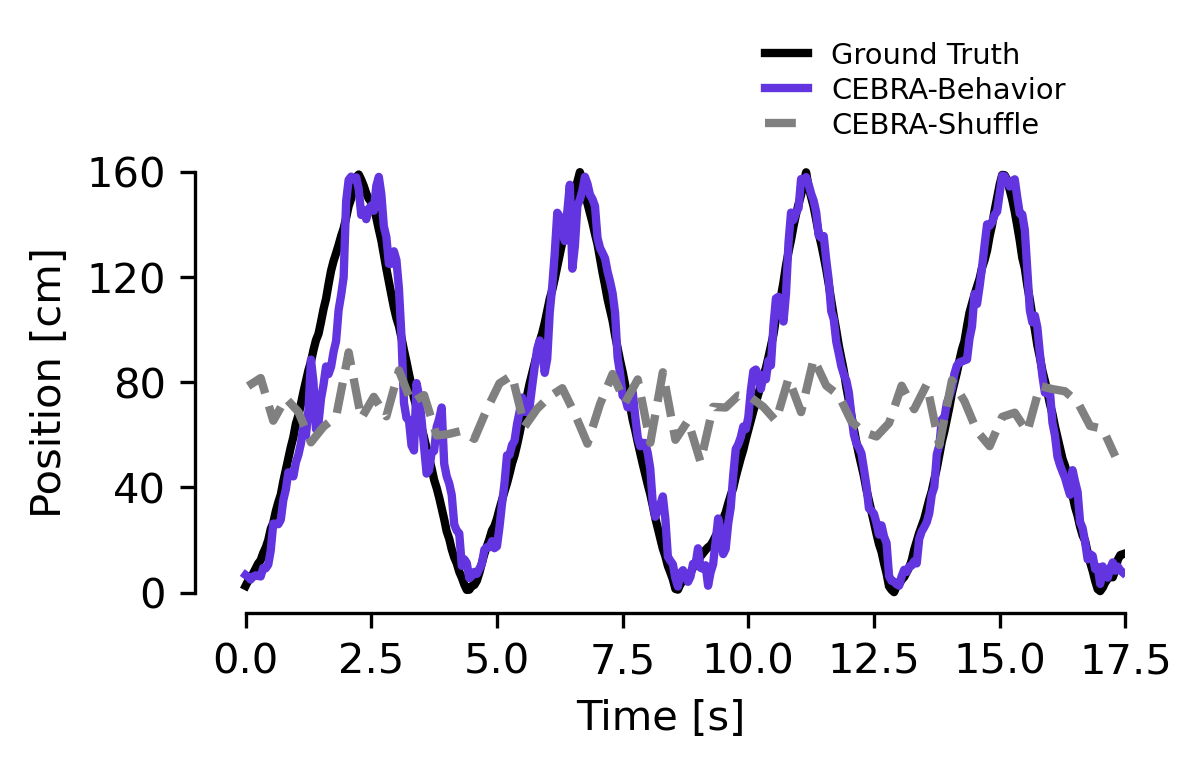
We utilized the hypothesis-driven (position) or the shuffle (erroneous) to decode the position of the rat, which produces a large difference in decoding performance: position+direction \(R^2\) is 73.35% vs. -49.90% shuffled and median absolute error 5.8 cm vs 44.7 cm. Purple line is decoding from the hypothesis-based latent space, dashed line is shuffled.
[6]:
# select timesteps
start_idx = 320
length = 700
history_len = 700
# load data
fs = data_fig_2d["embedding_all"].item()
test_fs = data_fig_2d["embedding_test"].item()
labels = data_fig_2d["true_all"].item()
test_labels = data_fig_2d["true_test"].item()
pred = data_fig_2d["prediction"].item()
pred_shuffle = data_fig_2d["prediction_shuffled"].item()
# plot
fig = plt.figure(figsize=(4, 2), dpi=300)
linewidth = 2
ax1_traj = plt.gca()
framerate = 25 / 1000
true_trajectory = ax1_traj.plot(
framerate * np.arange(length, step=2),
test_labels[start_idx : start_idx + length, 0][np.arange(length, step=2)] * 100,
"-",
c="k",
label="Ground Truth",
linewidth=linewidth,
)
(pred_trajectory,) = ax1_traj.plot(
framerate * np.arange(length, step=2),
pred[start_idx : start_idx + length, 0][np.arange(length, step=2)] * 100,
c="#6235E0",
label="CEBRA-Behavior",
linewidth=linewidth,
)
(pred_shuffle_trajectory,) = ax1_traj.plot(
framerate * (np.arange(length, step=10) + 2),
pred_shuffle[start_idx : start_idx + length, 0][np.arange(length, step=10)] * 100,
"--",
c="gray",
label="CEBRA-Shuffle",
linewidth=linewidth,
)
ax1_traj.set_yticks(np.linspace(0, 160, 5))
ax1_traj.spines["right"].set_visible(False)
ax1_traj.spines["top"].set_visible(False)
legend = ax1_traj.legend(
loc=(0.6, 1.0),
frameon=False,
handlelength=1.5,
labelspacing=0.25,
fontsize="x-small",
)
ax1_traj.set_xlabel("Time [s]")
ax1_traj.set_ylabel("Position [cm]")
ax1_traj.set_xlim([-1, 17.5])
ax1_traj.set_xticks(np.linspace(0, 17.5, 8))
ax1_traj.spines["bottom"].set_bounds(0, 17.5)
ax1_traj.spines["left"].set_bounds(0, 160)
plt.savefig("figure_2d_lines.svg", bbox_inches="tight", transparent=True)
plt.show()
Here is the performance across additional methods (The orange line indicates the median of the individual runs (n=10) that are indicated by black circles. Each run is averaged over 3 splits of the dataset).
[7]:
from matplotlib.markers import MarkerStyle
import warnings
import typing
import seaborn as sns
import matplotlib.pyplot as plt
ROOT = pathlib.Path("../data")
def recover_python_datatypes(element):
if isinstance(element, str):
if element.startswith("[") and element.endswith("]"):
if "," in element:
element = np.fromstring(element[1:-1], dtype=float, sep=",")
else:
element = np.fromstring(element[1:-1], dtype=float, sep=" ")
return element
def load_results(result_name):
"""Load a result file.
The first line in the result files specify the index columns,
the following lines are a CSV formatted file containing the
numerical results.
"""
results = {}
for result_csv in (ROOT / result_name).glob("*.csv"):
with open(result_csv) as fh:
index_names = fh.readline().strip().split(",")
df = pd.read_csv(fh).set_index(index_names)
df = df.applymap(recover_python_datatypes)
results[result_csv.stem] = df
return results
def show_boxplot(df, metric, ax, labels=None, color="C1"):
with warnings.catch_warnings():
warnings.simplefilter("ignore")
sns.boxplot(
data=df,
y="method",
x=metric,
orient="h",
order=labels,
width=0.5,
color="k",
linewidth=2,
flierprops=dict(alpha=0.5, markersize=0, marker=".", linewidth=0),
medianprops=dict(
c=color, markersize=0, marker=".", linewidth=2, solid_capstyle="round"
),
whiskerprops=dict(solid_capstyle="butt", linewidth=0),
showbox=False,
showcaps=False,
ax=ax,
)
marker_style = MarkerStyle("o", "none")
sns.stripplot(
data=df,
y="method",
x=metric,
orient="h",
size=4,
color="k",
order=labels,
marker=marker_style,
linewidth=1,
ax=ax,
alpha=0.75,
jitter=0.15,
zorder=-1,
)
ax.set_ylabel("")
sns.despine(left=True, bottom=False, ax=ax)
ax.tick_params(
axis="x", which="both", bottom=True, top=False, length=5, labelbottom=True
)
return ax
def _add_value(df, **kwargs):
for key, value in kwargs.items():
df[key] = value
return df
def join(results):
return pd.concat([_add_value(df, method=key) for key, df in results.items()])
def get_metrics(results):
for key, results_ in results.items():
df = results_.copy()
df["method"] = key
df["test_position_error"] *= 100
df = df[df.animal == 0].pivot_table(
"test_position_error", index=("method", "seed"), aggfunc="mean"
)
yield df
[8]:
autolfads = pd.read_csv("../data/autolfads_decoding_2d_full.csv", index_col=0)
autolfads = autolfads.rename(columns={"split": "repeat", "rat": "animal"})
autolfads["animal"] = autolfads["animal"].apply(lambda v: "abcg".index(v[0]))
[9]:
results = load_results(result_name="results_v1")
results["pivae-mc"] = pd.read_csv("../data/Figure2/figure2_pivae_mcmc.csv", index_col=0)
results["autolfads"] = autolfads
df = pd.concat(get_metrics(results)).reset_index()
plt.figure(figsize=(2, 2), dpi=200)
ax = plt.gca()
show_boxplot(
df=df,
metric="test_position_error",
ax=ax,
color="C1",
labels=[
"cebra-b",
"pivae-mc",
"pivae-wo",
"cebra-t",
"autolfads",
"tsne",
"umap",
"pca",
],
)
ticks = [0, 10, 20, 30, 40]
ax.set_xlim(min(ticks), max(ticks))
ax.set_xticks(ticks)
ax.set_xlabel("Error [cm]")
ax.set_yticklabels(
[
"CEBRA-Behavior",
"conv-pi-VAE (MC)",
"conv-piVAE (kNN)",
"CEBRA-Time",
"autoLFADS",
"t-SNE",
"UMAP",
"PCA",
]
)
# plt.savefig("figure2d.svg", bbox_inches = "tight", transparent = True)
plt.show()
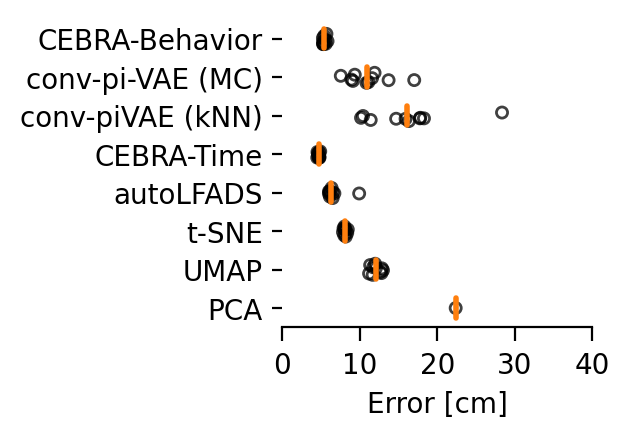
Figure 2f#
Left: Visualization of the neural embeddings computed with different input dimensions, and the related persistent co-homology lifespan diagrams below.
[10]:
topology_viz = data["topology"]["viz"]
fig = plt.figure(figsize=(15, 5))
for i, dim in enumerate([3, 8, 16]):
ax = fig.add_subplot(1, 3, i + 1, projection="3d")
emb = topology_viz[dim]
label = topology_viz["label"]
r = label[:, 1] == 1
l = label[:, 2] == 1
idx1, idx2, idx3 = (0, 1, 2)
if i == 1:
idx1, idx2, idx3 = (5, 6, 7)
ax.scatter(
emb[l, idx1], emb[l, idx2], emb[l, idx3], c=label[l, 0], cmap="cool", s=0.1
)
ax.scatter(
emb[r, idx1], emb[r, idx2], emb[r, idx3], cmap="viridis", c=label[r, 0], s=0.1
)
ax.axis("off")
ax.set_title(f"Dimension {dim}", fontsize=20)
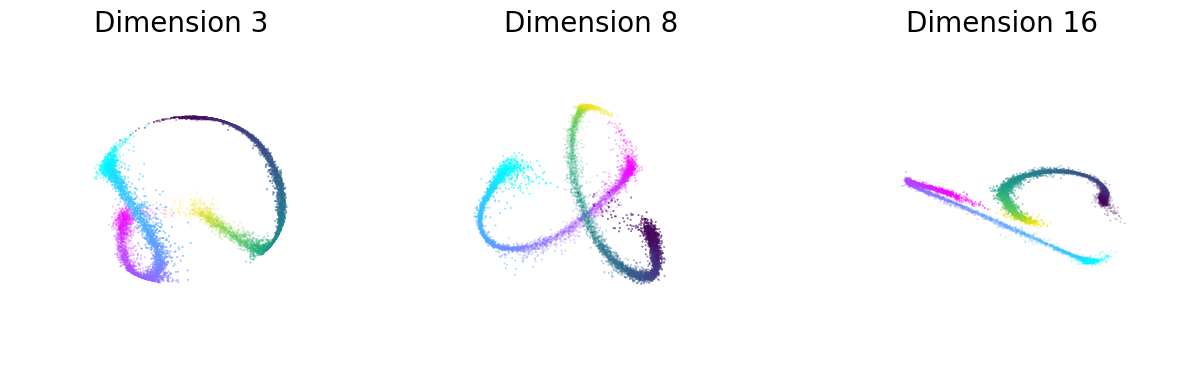
Figure 2f#
Right: Betti numbers from shuffled embeddings (Sh.) and across increasing dimensions.
[11]:
dims = [3, 8, 16]
cocycle = ["Points", "Loops", "Voids"]
colors = ["b", "orange", "green"]
for d in range(3):
topology_result = data["topology"]["behavior_topology"][dims[d]][
"dgms"
] # analysis_offsets[d]['dgms']
fig, axs = plt.subplots(3, 1, sharex=True, figsize=(7, 8))
fig.suptitle(f"Dimension{dims[d]}", fontsize=30)
axs[0].set_xlim(0, 2)
for k in range(3):
bars = topology_result[k]
bars[bars == np.inf] = 10
lc = (
np.vstack(
[
bars[:, 0],
np.arange(len(bars), dtype=int) * 6,
bars[:, 1],
np.arange(len(bars), dtype=int) * 6,
]
)
.swapaxes(1, 0)
.reshape(-1, 2, 2)
)
line_segments = LineCollection(lc, linewidth=5, color="gray", alpha=0.5)
axs[k].set_ylabel(cocycle[k], fontsize=20)
if k == 0:
axs[k].set_ylim(len(bars) * 6 - 120, len(bars) * 6)
elif k == 1:
axs[k].set_ylim(0, len(bars) * 1 - 30)
elif k == 2:
axs[k].set_ylim(0, len(bars) * 6 + 10)
axs[k].add_collection(line_segments)
axs[k].set_yticks([])
if k == 2:
axs[k].set_xticks(np.linspace(0, 2, 3), np.linspace(0, 2, 3), fontsize=15)
axs[k].set_xlabel("Lifespan", fontsize=20)
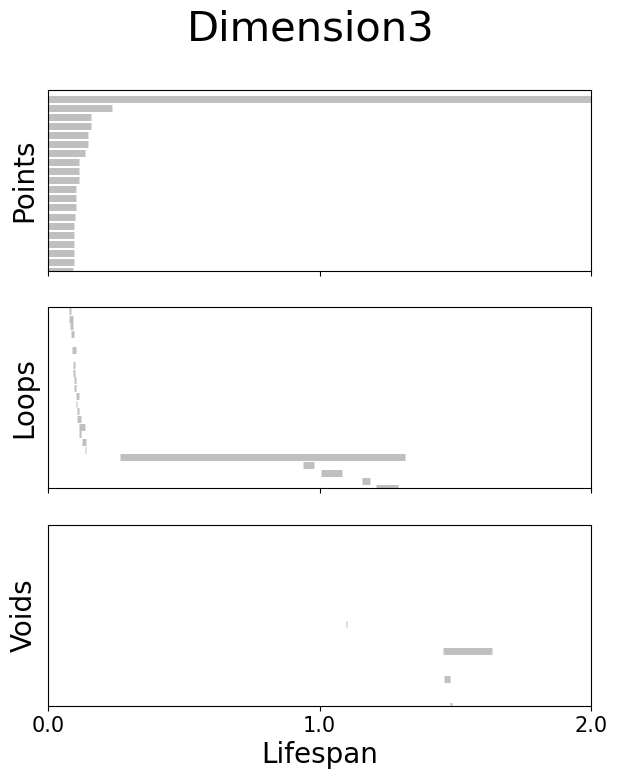
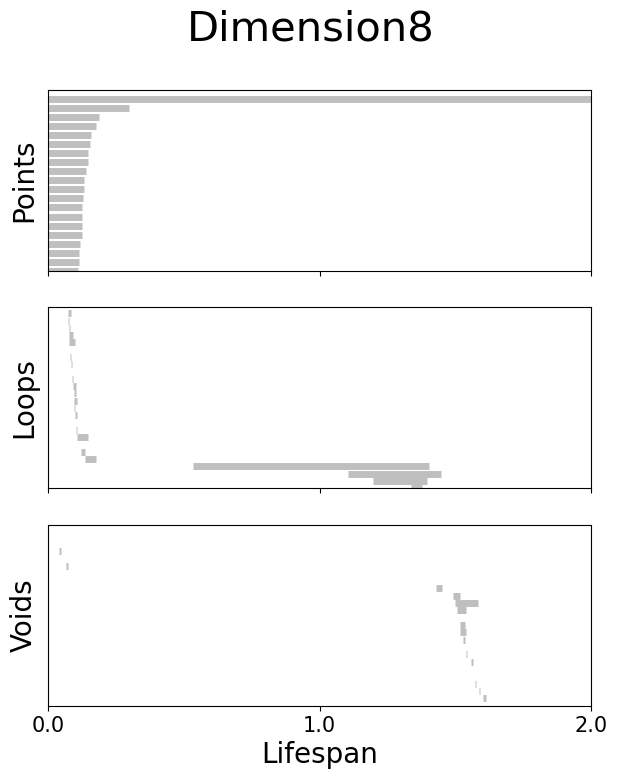
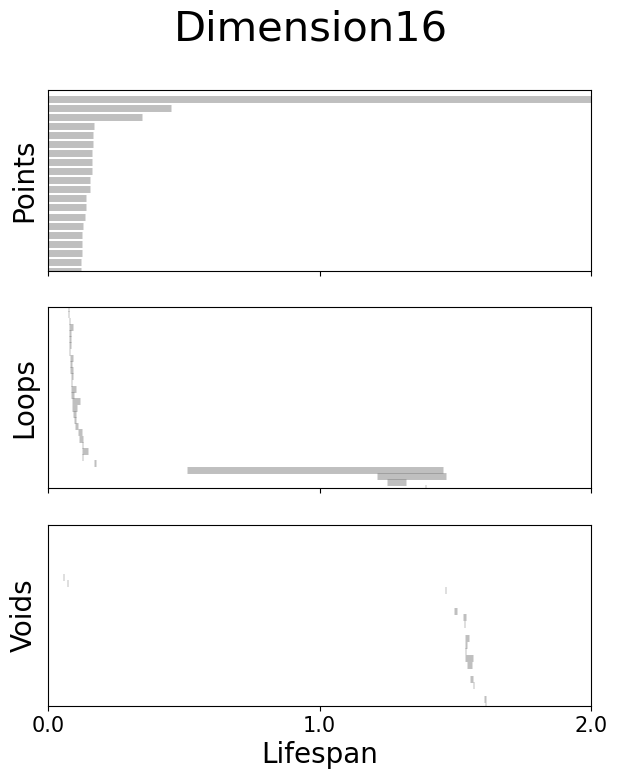
Figure 2g#
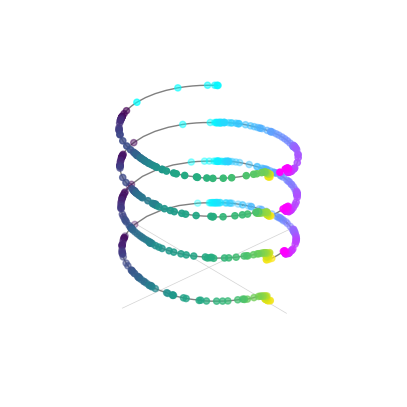
Topology preserving circular coordinates using the first co-cycle from persistent co-homology analysis
[12]:
circular_coord = data["topology"]["circular_coord"]
fig = plt.figure(figsize=(5, 7))
ax = plt.subplot(projection="3d", computed_zorder=False)
angle = np.linspace(0, 21, 300)
label = circular_coord["label"]
radial_angle = circular_coord["radial_angle"]
r_ind = label[:, 1] == 1
l_ind = label[:, 2] == 1
x = np.cos(radial_angle)
y = np.sin(radial_angle)
z = np.cumsum(label[:, 0])
fine_angle = circular_coord["finer_angle"]
fine_z = circular_coord["fine_z"]
ax.scatter(x[r_ind], y[r_ind], z[r_ind], c=label[r_ind, 0], cmap="cool", zorder=-1)
ax.scatter(x[l_ind], y[l_ind], z[l_ind], c=label[l_ind, 0], cmap="viridis", zorder=-1)
ax.plot(np.cos(fine_angle), np.sin(fine_angle), fine_z, c="gray", zorder=-10, lw=1)
radius = 1.3
offset = 180
ax.plot(
[
radius * np.cos(radial_angle[offset]),
radius * np.cos(np.pi + radial_angle[offset]),
],
[
radius * np.sin(radial_angle[offset]),
radius * np.sin(np.pi + radial_angle[offset]),
],
[0, 0],
lw=0.5,
c="lightgray",
zorder=-20,
)
ax.plot(
[
radius * np.cos(radial_angle[offset] + np.pi / 2),
radius * np.cos(radial_angle[offset] + np.pi * 3 / 2),
],
[
radius * np.sin(radial_angle[offset] + np.pi / 2),
radius * np.sin(3 / 2 * np.pi + radial_angle[offset]),
],
[0, 0],
lw=0.5,
c="lightgray",
zorder=-20,
)
ax.axis("off")
[12]:
(-1.3577543976255955,
1.3577543976255955,
-1.3577543976255955,
1.3577543976255955)